Ahmedshehata (talk | contribs) m (Ahmedshehata moved page Draft Hagag 877846790 to Review 482003212394) |
|||
| (50 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
american<math>^{2,*}</math>E-mail:ahmed.shehata@su.edu.eg''' | american<math>^{2,*}</math>E-mail:ahmed.shehata@su.edu.eg''' | ||
--> | --> | ||
| − | |||
==Abstract== | ==Abstract== | ||
Food security has become a significant issue due to the growing human population. In this case, a significant role is played by agriculture. The essential foods are obtained mainly from plants. Plant diseases can, however, decrease both food production and its quality. Therefore, it is substantial to comprehend the dynamics of plant diseases as they can provide insightful information about the dispersal of plant diseases. In order to investigate the dynamics of plant disease and analyze the effects of strategies of disease control, a mathematical model can be applied. We show that this model provides the non-negative solutions that population dynamics requires. The model was investigated by using the Atangana-Baleanu in Caputo sense (ABC) operator which is symmetrical to the Caputo-Fabrizio (CF) operator with a different function. Whereas the ABC operator uses the generalized Mittag-Leffler function while the CF operator employs the exponential kernel. For the proposed model, we have displayed the local and global stability of a nonendemic and an endemic equilibrium, existence and uniqueness theorems. By applying the fractional Adams-Bashforth-Moulton method, we have implemented numerical solutions to illustrate the theoretical analysis. | Food security has become a significant issue due to the growing human population. In this case, a significant role is played by agriculture. The essential foods are obtained mainly from plants. Plant diseases can, however, decrease both food production and its quality. Therefore, it is substantial to comprehend the dynamics of plant diseases as they can provide insightful information about the dispersal of plant diseases. In order to investigate the dynamics of plant disease and analyze the effects of strategies of disease control, a mathematical model can be applied. We show that this model provides the non-negative solutions that population dynamics requires. The model was investigated by using the Atangana-Baleanu in Caputo sense (ABC) operator which is symmetrical to the Caputo-Fabrizio (CF) operator with a different function. Whereas the ABC operator uses the generalized Mittag-Leffler function while the CF operator employs the exponential kernel. For the proposed model, we have displayed the local and global stability of a nonendemic and an endemic equilibrium, existence and uniqueness theorems. By applying the fractional Adams-Bashforth-Moulton method, we have implemented numerical solutions to illustrate the theoretical analysis. | ||
| − | '''Keywords | + | '''Keywords''': Adams-Bashforth-Moulton method, local and global asymptotic equilibrium stability, sensitivity analysis, existence theorems, uniqueness theorems, plant diseases model, numerical simulations |
| − | ==1 Introduction== | + | ==1. Introduction== |
Plants are an incredibly valuable component of our world. The earth, due to the presence of plants, is known as a green planet. They are perhaps the most important component of the life of all the earth's living beings. Some of the plant's essential functions are food, reducing the level of pollution, supplying fresh oxygen, medications, furniture and refuge. Plant disease, however, triggers a decline in food production and efficiency, which can lead to numerous health and social issues. Moreover, it can cause considerable economic ramifications. | Plants are an incredibly valuable component of our world. The earth, due to the presence of plants, is known as a green planet. They are perhaps the most important component of the life of all the earth's living beings. Some of the plant's essential functions are food, reducing the level of pollution, supplying fresh oxygen, medications, furniture and refuge. Plant disease, however, triggers a decline in food production and efficiency, which can lead to numerous health and social issues. Moreover, it can cause considerable economic ramifications. | ||
| − | Plant disease epidemiology studies the evolution of populations of plant diseases in time and space. Usually, the execution of the techniques is accomplished by roguing and then replanting, which offers two possible advantages. Initially, infected plants could be removed and could inoculum sources could be reduced, possibly slowing the dispersal of pathogens. Then, the infected plant may die or suffer a reduction in yield. Consequently, substituting diseased plants with healthful plants can indemnity crop casualties. The exposed plant can also be able to spread disease in some cases [1 | + | Plant disease epidemiology studies the evolution of populations of plant diseases in time and space. Usually, the execution of the techniques is accomplished by roguing and then replanting, which offers two possible advantages. Initially, infected plants could be removed and could inoculum sources could be reduced, possibly slowing the dispersal of pathogens. Then, the infected plant may die or suffer a reduction in yield. Consequently, substituting diseased plants with healthful plants can indemnity crop casualties. The exposed plant can also be able to spread disease in some cases [1-3]. Since the exposed plant is capable of spreading disease, the propagation of plant disease can be more rapid. It is therefore important to realize the impact of plants uncovered on the dynamics of plant disease contamination. Their simulation revealed that the application of fungicides is efficient in minimizing population infection [4-6]. |
| + | |||
| + | Mathematical modeling is useful in explaining how diseases dispersal and different factors involved in the dispersal of the disease have been specified [7-11]. The defensive and curative fungicide model was introduced in Anggriani et al. [12], where it has been split into three ingredients: Infectious, protected and susceptible. Their simulation revealed that the implementation of fungicides is active in minimizing population infection. Model plant diseases with replanting, roguing and preventive care have been introduced in Anggriani et al. [13] without consideration to curative therapy. In 2017, Anggriani et al. [14] created a plant disease mathematical model that includes five ingredients: Susceptible, Protected, Infectious, Exposed and Post-Infectious with protective curative therapies. They observed that by using curative and preventative care, the transmission of plant disease can be minimized. However, where only one therapy is offered, preventive therapy is favored over curative therapy. | ||
| + | |||
| + | In actuality, there are various meanings of fractional derivatives which in general do not necessarily correspond. One of these definitions which is often utilized in different implementations of fractional differential equations is Caputo fractional derivative [15-18]. There is also a novel fractional derivative definition, named the Caputo-Fabrizio fractional operator, which centered on the exponential function [19-21]. And in the same context, there is a novel fractional derivative definition, named the Atangana-Baleanu fractional operator, which centered on the Mittag-Leffler function [22-27]. Many authors have successfully attempted to model actual processes utilizing this fractional derivative operator [28-29]. | ||
| + | |||
| + | We consider a plant disease spread model within fractional calculus, where a susceptible person crosses an exposed stage prior to becoming an infectious person and diseases may also be passed on by exposed plants. The major purpose for this protraction is that the plant diseases of the classical case [12-14] do not load any acquaintance about the memory and learning techniques that impact the propagation of a disease [25]. Now, we regard the plant diseases model with fractional order as follows: | ||
<span id="eq-1"></span> | <span id="eq-1"></span> | ||
| Line 33: | Line 38: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }S_{1}\left(t\right) = r(M-N)-\gamma S_{1}\left(t\right)-\frac{a_{1}}{M}S_{1}\left(t\right)I_{1}\left(t\right)-\alpha S_{1}\left(t\right)+\beta P_{1}\left(t\right), | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }S_{1}\left(t\right) & = r(M-N)-\gamma S_{1}\left(t\right)-\frac{a_{1}}{M}S_{1}\left(t\right)I_{1}\left(t\right)-\alpha S_{1}\left(t\right)+\beta P_{1}\left(t\right),\\ |
| − | + | ^{ABC}D_{*}^{\upsilon }P_{1}\left(t\right) & = \alpha S_{1}\left(t\right)+\beta P_{1}\left(t\right)-\gamma P_{1}\left(t\right),\\ | |
| − | + | ^{ABC}D_{*}^{\upsilon }E_{1}\left(t\right) & =\frac{a_{1}}{M}S_{1}\left(t\right)I_{1}\left(t\right) -\left(\gamma + a_{2}+b_{1}\right) E_{1}\left(t\right),\\ | |
| − | + | ^{ABC}D_{*}^{\upsilon }I_{1}\left(t\right) & = a_{2}E_{1}\left(t\right)-\left(\gamma{+}a_{3}+b_{2}+\eta \right)I_{1}\left(t\right),\\ | |
| − | + | ^{ABC}D_{*}^{\upsilon }R_{1}\left(t\right) &= a_{3}I_{1}\left(t\right)-\left(\gamma{+}b_{3}\right)R_{1}\left(t\right). \end{align}</math> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
|} | |} | ||
| + | | style="width: 5px;text-align: right;white-space: nowrap;" |(1) | ||
|} | |} | ||
With initial conditions <math>\left(S_{1}\right)_{0}\left(0\right)>0</math>, <math>\left(P_{1}\right)_{0}\left(0\right)\geq{0}</math>, <math>\left(E_{1}\right)_{0}\left(0\right)\geq{0}</math>, <math>\left(I_{1}\right)_{0}\left(0\right)\geq{0}</math> and <math>\left(R_{1}\right)_{0}\left(0\right)\geq{0}</math>, where <math>N</math> indicate to the overall population of the actual plant, <math>S_{1}\left(t\right)</math> is representing the Susceptible population, <math>P_{1}\left(t\right)</math> is representing the Protected population, <math>E_{1}\left(t\right)</math> is representing the Exposed population, <math>I_{1}\left(t\right)</math> is representing the Infectious/Removed population, <math>R_{1}\left(t\right)</math> is representing the Recovered population, and <math>N=S_{1}\left(t\right)+P_{1}\left(t\right)+E_{1}\left(t\right)+I_{1}\left(t\right)+R_{1}\left(t\right)</math>, this suggests that the size of population is not steady. (i.e. size of the population is variable). The actual meaning of each of the model's parameters, all of which have positive values, is as follows: | With initial conditions <math>\left(S_{1}\right)_{0}\left(0\right)>0</math>, <math>\left(P_{1}\right)_{0}\left(0\right)\geq{0}</math>, <math>\left(E_{1}\right)_{0}\left(0\right)\geq{0}</math>, <math>\left(I_{1}\right)_{0}\left(0\right)\geq{0}</math> and <math>\left(R_{1}\right)_{0}\left(0\right)\geq{0}</math>, where <math>N</math> indicate to the overall population of the actual plant, <math>S_{1}\left(t\right)</math> is representing the Susceptible population, <math>P_{1}\left(t\right)</math> is representing the Protected population, <math>E_{1}\left(t\right)</math> is representing the Exposed population, <math>I_{1}\left(t\right)</math> is representing the Infectious/Removed population, <math>R_{1}\left(t\right)</math> is representing the Recovered population, and <math>N=S_{1}\left(t\right)+P_{1}\left(t\right)+E_{1}\left(t\right)+I_{1}\left(t\right)+R_{1}\left(t\right)</math>, this suggests that the size of population is not steady. (i.e. size of the population is variable). The actual meaning of each of the model's parameters, all of which have positive values, is as follows: | ||
| − | * <math display="inline">r</math> | + | * <math display="inline">r</math>: rate of replanting. |
| − | * <math display="inline">a_{1}</math> disease progression diversion rate for latent compartment. | + | * <math display="inline">a_{1}</math>: disease progression diversion rate for latent compartment. |
| − | * <math display="inline">a_{2}</math> disease progression diversion rate for infected compartment. | + | * <math display="inline">a_{2}</math>: disease progression diversion rate for infected compartment. |
| − | * <math display="inline">a_{3}</math> disease progression diversion rate for removed compartment. | + | * <math display="inline">a_{3}</math>: disease progression diversion rate for removed compartment. |
| − | * <math display="inline">M</math> overall maximum plant population (agronomy). | + | * <math display="inline">M</math>: overall maximum plant population (agronomy). |
| − | * <math display="inline">b_{1}</math> influence accumulative death rate for latent compartment. | + | * <math display="inline">b_{1}</math>: influence accumulative death rate for latent compartment. |
| − | * <math display="inline">b_{2}</math> influence accumulative death rate for infected compartment. | + | * <math display="inline">b_{2}</math>: influence accumulative death rate for infected compartment. |
| − | * <math display="inline">b_{3}</math> influence accumulative death rate for latent removed compartment. | + | * <math display="inline">b_{3}</math>: influence accumulative death rate for latent removed compartment. |
| − | * <math display="inline">\alpha </math> efficacy preventive therapy. | + | * <math display="inline">\alpha </math>: efficacy preventive therapy. |
| − | * <math display="inline">\beta </math> preventive therapy rate. | + | * <math display="inline">\beta </math>: preventive therapy rate. |
| − | * <math display="inline">\gamma </math> rate of natural death. | + | * <math display="inline">\gamma </math>: rate of natural death. |
| − | * <math display="inline">\eta </math> rate of roguing. | + | * <math display="inline">\eta </math>: rate of roguing. |
| + | |||
Some assumptions have been made to facilitate understanding this model [american14]: | Some assumptions have been made to facilitate understanding this model [american14]: | ||
* There is no closure of the plant population due to replanting and natural death. | * There is no closure of the plant population due to replanting and natural death. | ||
| − | * The infected plant compartment comprises of two compartments, called, latent E(t) and infected I(t). | + | * The infected plant compartment comprises of two compartments, called, latent <math display="inline">E(t)</math> and infected <math display="inline">I(t)</math>. |
* The infected plant is lifted if it shows comprehensive symptoms. | * The infected plant is lifted if it shows comprehensive symptoms. | ||
* The insect vector and environment factor are neglected. | * The insect vector and environment factor are neglected. | ||
| Line 71: | Line 73: | ||
* Protected plants have defensive or prevention impact, but are not immune to disease, therefore it is permitting re-entry into the susceptible compartment. | * Protected plants have defensive or prevention impact, but are not immune to disease, therefore it is permitting re-entry into the susceptible compartment. | ||
| − | |||
| − | + | This article is structured to have some significant preliminaries in Section 2. Section 3 transacts with studying the local and global asymptotic equilibrium stability (the disease-free case and the endemic case) and the sensibility analysis of reproduction number (<math display="inline">\mathrm{\mathcal{R}}_{0}</math>) without control of our model (Eq.[[#eq-1|(1)]]). Section 4 transacts with studying the existence and uniqueness theorems of our model (Eq.[[#eq-1|(1)]]). Then, computational technique (Adams-Bashforth-Moulton method) are graphically represented and covered in Sections 5 and 6. Finally, conclusions are drawn. | |
| − | Recently, many fractional calculus concepts and definitions have been developed [ | + | ==2. Preliminaries== |
| + | |||
| + | Recently, many fractional calculus concepts and definitions have been developed [31-32]. | ||
===Definition 1=== | ===Definition 1=== | ||
| Line 112: | Line 115: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>_{0}^{AB}J_{\varrho }^{\upsilon }\varphi \left(\varrho \right) = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\varphi \left(\varrho \right)+\frac{\upsilon }{\chi \left(\upsilon \right)\Gamma \left(\upsilon \right)}\stackrel=0\varphi \left(\tau \right)\left(\varrho{-\tau}\right)^{\upsilon{-1}}d\tau , | + | | style="text-align: center;" | <math>\begin{align} _{0}^{AB}J_{\varrho }^{\upsilon }\varphi \left(\varrho \right) & = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\varphi \left(\varrho \right)+\frac{\upsilon }{\chi \left(\upsilon \right)\Gamma \left(\upsilon \right)}\stackrel=0\varphi \left(\tau \right)\left(\varrho{-\tau}\right)^{\upsilon{-1}}d\tau ,\\ |
| − | + | & = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\varphi \left(\varrho \right)+\frac{\upsilon }{\chi \left(\upsilon \right)\Gamma \left(\upsilon \right)}\left(I^{\upsilon }\varphi \left(\varrho \right)\right).\end{align} </math> | |
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (4) | | style="width: 5px;text-align: right;white-space: nowrap;" | (4) | ||
|} | |} | ||
| − | ==3 Analysis of plant disease model== | + | ==3. Analysis of plant disease model== |
===3.1 The local asymptotic equilibrium stability=== | ===3.1 The local asymptotic equilibrium stability=== | ||
| − | Since <math display="inline">^{ABC}D^{\upsilon }S_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }P_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }E_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }I_{1}\left(t\right)=0</math> and <math display="inline">^{ABC}D^{\upsilon }R_{1}\left(t\right)=0</math> when <math display="inline">t\rightarrow \infty </math>, we can use them to find two equilibrium points of the fractional system ([[#eq-1|1]]), by solving the above equations we get | + | Since <math display="inline">^{ABC}D^{\upsilon }S_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }P_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }E_{1}\left(t\right)=0</math>, <math display="inline">^{ABC}D^{\upsilon }I_{1}\left(t\right)=0</math> and <math display="inline">^{ABC}D^{\upsilon }R_{1}\left(t\right)=0</math> when <math display="inline">t\rightarrow \infty </math>, we can use them to find two equilibrium points of the fractional system (Eq.[[#eq-1|(1)]]), by solving the above equations we get |
| − | ====3.1.1 Non endemic equilibrium (NEE) point | + | ====3.1.1 Non endemic equilibrium (NEE) point==== |
| − | The NEE solution of the system ([[#eq-1|1]]) is | + | The NEE solution of the system (Eq.[[#eq-1|(1)]]) is |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 134: | Line 136: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\mathcal{E}_{0}=\left(\left(S_{1}\right)_{eq},\left(P_{1}\right)_{eq},\left(E_{1}\right)_{eq},\left(I_{1}\right)_{eq},\left(R_{1}\right)_{eq}\right)=\left(\frac{\gamma A_{3}Mr}{(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))},-\frac{\alpha \gamma Mr}{(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))},0,0,0\right), </math> | + | | style="text-align: center;" | <math>\begin{align} \mathcal{E}_{0} & =\left(\left(S_{1}\right)_{eq},\left(P_{1}\right)_{eq},\left(E_{1}\right)_{eq},\left(I_{1}\right)_{eq},\left(R_{1}\right)_{eq}\right)\\ |
| + | & =\left(\frac{\gamma A_{3}Mr}{(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))},-\frac{\alpha \gamma Mr}{(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))},0,0,0\right), \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (5) | | style="width: 5px;text-align: right;white-space: nowrap;" | (5) | ||
| Line 151: | Line 154: | ||
|} | |} | ||
| − | It is known as the number of secondary contagions induced by a single primary contagion in a completely susceptible population [ | + | It is known as the number of secondary contagions induced by a single primary contagion in a completely susceptible population [35] and is generally expressed using model (Eq.[[#eq-1|(1)]]). The rate of the basic reproduction number counts on the replanting rate value. This is the justification that we should replant the plant if we want to control plant disease. |
| − | ====3.1.2 | + | ====3.1.2 Endemic equilibrium (EE) point==== |
| + | |||
| + | The endemic equilibrium solution of the system (Eq.[[#eq-1|(1)]]) is | ||
| + | |||
| + | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| + | |- | ||
| + | | | ||
| + | {| style="text-align: left; margin:auto;width: 100%;" | ||
| + | |- | ||
| + | | style="text-align: center;" |<math>\mathcal{E}^{*}=\left(S_{1}^{*},P_{1}^{*},E_{1}^{*},I_{1}^{*},R_{1}^{*}\right),</math> | ||
| + | |} | ||
| + | |} | ||
where | where | ||
| Line 162: | Line 176: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\begin{cases}S_{1}^{*}= & \frac{A_{1}A_{2}M}{a_{1}a_{2}},\\ P_{1}^{*}= & -\frac{\alpha A_{1}A_{2}M}{a_{1}a_{2}A_{3}},\\ E_{1}^{*}= & \frac{A_{2}M\left(b_{3}+\gamma \right)\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}a_{2}A_{3}\left(a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)-A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)\right)},\\ I_{1}^{*}= & -\frac{M\left(b_{3}+\gamma \right)\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}A_{3}\left(A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)-a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)\right)},\\ R_{1}^{*}= & -\frac{a_{3}M\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}A_{3}\left(A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)-a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)\right)}. \end{cases} </math> | + | | style="text-align: center;" | <math>\begin{cases}S_{1}^{*}= & \displaystyle\frac{A_{1}A_{2}M}{a_{1}a_{2}},\\ P_{1}^{*}= & -\displaystyle\frac{\alpha A_{1}A_{2}M}{a_{1}a_{2}A_{3}},\\ E_{1}^{*}= & \displaystyle\frac{A_{2}M\left(b_{3}+\gamma \right)\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}a_{2}A_{3}\left(a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)-A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)\right)},\\ I_{1}^{*}= & -\displaystyle\frac{M\left(b_{3}+\gamma \right)\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}A_{3}\left(A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)-a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)\right)},\\ R_{1}^{*}= & -\displaystyle\frac{a_{3}M\left(A_{1}A_{2}(\gamma{+}r)(\alpha \beta{+}A_{3}(\alpha{+\gamma}))-a_{1}a_{2}\gamma A_{3}r\right)}{a_{1}A_{3}\left(A_{2}\left(b_{3}+\gamma \right)\left(A_{1}(\gamma{+}r)-b_{1}r\right)-a_{2}r\left(b_{3}\left(a_{3}+b_{2}+\eta \right)+\gamma \left(b_{2}+\eta \right)\right)\right)}. \end{cases} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (7) | | style="width: 5px;text-align: right;white-space: nowrap;" | (7) | ||
|} | |} | ||
| − | with <math display="inline">N^{*}=\frac{rM-b_{1}E_{1}^{*}-\left(b_{2}+\eta \right)I_{1}^{*}-b_{3}R_{1}^{*}}{\gamma{+}r}</math>. Conclusion of the outcomes of the system's equilibrium points ([[#eq-1|1]]) with prepared to research analytically the stability of the equilibrium points. | + | with <math display="inline">N^{*}=\displaystyle\frac{rM-b_{1}E_{1}^{*}-\left(b_{2}+\eta \right)I_{1}^{*}-b_{3}R_{1}^{*}}{\gamma{+}r}</math>. Conclusion of the outcomes of the system's equilibrium points (Eq.[[#eq-1|(1)]]) with prepared to research analytically the stability of the equilibrium points. |
| − | ''' | + | '''Theorem 3.1.2.1'''. The NEE point <math display="inline">\mathcal{E}_{0}</math> of model (Eq.[[#eq-1|(1)]]) is locally asymptotically stable if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and unstable if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>. |
| − | '''Proof''' The system's Jacobian matrix ([[#eq-1|1]]) at <math>\mathcal{E}_{0}</math> is | + | '''Proof'''. The system's Jacobian matrix (Eq.[[#eq-1|(1)]]) at <math>\mathcal{E}_{0}</math> is |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 178: | Line 192: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>J_{\mathcal{E}_{0}}=\left(\begin{array}{ccccc}-\left(\alpha{+\gamma}\right)& \beta & 0 & \frac{ra_{1}\left(\beta{+\gamma}\right)}{\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ \alpha & -\left(\beta{+\gamma}\right)& 0 & 0 & 0\\ 0 & 0 & -A_{1} & \frac{ra_{1}\left(\beta{+\gamma}\right)}{\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ 0 & 0 & a_{2} & -A_{2} & 0\\ 0 & 0 & 0 & a_{3} & -\left(\gamma{+}b_{3}\right) \end{array}\right). </math> | + | | style="text-align: center;" | <math>J_{\mathcal{E}_{0}}=\left(\begin{array}{ccccc}-\left(\alpha{+\gamma}\right)& \beta & 0 & \displaystyle\frac{ra_{1}\left(\beta{+\gamma}\right)}{\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ \alpha & -\left(\beta{+\gamma}\right)& 0 & 0 & 0\\ 0 & 0 & -A_{1} & \displaystyle\frac{ra_{1}\left(\beta{+\gamma}\right)}{\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ 0 & 0 & a_{2} & -A_{2} & 0\\ 0 & 0 & 0 & a_{3} & -\left(\gamma{+}b_{3}\right) \end{array}\right). </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (8) | | style="width: 5px;text-align: right;white-space: nowrap;" | (8) | ||
| Line 196: | Line 210: | ||
|} | |} | ||
| − | From Eq. ([[#eq-9|9]]), we can notice that if <math display="inline">\mathrm{\mathcal{R}}_{0}\leq{1}</math>, then <math display="inline">\mathcal{E}_{0}</math> is locally asymptotically stable. This suggests that all polynomial coefficients ([[#eq-9|9]]) have the same signal, then the eigenvalues (roots) have negative real part. But if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, the NEE is unstable and this would lead to a stable endemic equilibrium being present of <math display="inline">\mathcal{E}^{*}</math>. Now by proving the theorem of local stability of <math display="inline">\mathcal{E}^{*}</math> , we conclude this section. | + | From Eq.([[#eq-9|9]]), we can notice that if <math display="inline">\mathrm{\mathcal{R}}_{0}\leq{1}</math>, then <math display="inline">\mathcal{E}_{0}</math> is locally asymptotically stable. This suggests that all polynomial coefficients ([[#eq-9|9]]) have the same signal, then the eigenvalues (roots) have negative real part. But if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, the NEE is unstable and this would lead to a stable endemic equilibrium being present of <math display="inline">\mathcal{E}^{*}</math>. Now by proving the theorem of local stability of <math display="inline">\mathcal{E}^{*}</math> , we conclude this section. |
| − | ''' | + | '''Theorem 3.1.2.2'''. If <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, the endemic equilibrium <math>\mathcal{E}^{*}</math> of model (Eq.[[#eq-1|(1)]]) is locally asymptotically stable. |
| − | '''Proof''' The system's Jacobian matrix ([[#eq-1|1]]) at <math display="inline">\mathcal{E}^{*}</math> is | + | '''Proof'''. The system's Jacobian matrix (Eq.[[#eq-1|(1)]]) at <math display="inline">\mathcal{E}^{*}</math> is |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 207: | Line 221: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>J_{\mathcal{E}^{*}}=\left(\begin{array}{ccccc}-\left(\beta{+\gamma}\right)-\frac{\gamma \left(\mathrm{\mathcal{R}}_{0}-1\right)\left(\alpha{+\beta}+\gamma \right)}{\left(\beta{+\gamma}\right)} & \beta & 0 & \frac{ra_{1}\left(\beta{+\gamma}\right)}{\mathrm{\mathcal{R}}_{0}\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ \alpha & -\left(\beta{+\gamma}\right)& 0 & 0 & 0\\ \frac{\gamma \left(\mathrm{\mathcal{R}}_{0}-1\right)\left(\alpha{+\beta}+\gamma \right)}{\left(\beta{+\gamma}\right)} & 0 & -A_{1} & \frac{ra_{1}\left(\beta{+\gamma}\right)}{\mathrm{\mathcal{R}}_{0}\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ 0 & 0 & a_{2} & -A_{2} & 0\\ 0 & 0 & 0 & a_{3} & -\left(\gamma{+}b_{3}\right) \end{array}\right), </math> | + | | style="text-align: center;" | <math>J_{\mathcal{E}^{*}}=\left(\begin{array}{ccccc}-\left(\beta{+\gamma}\right)-\displaystyle\frac{\gamma \left(\mathrm{\mathcal{R}}_{0}-1\right)\left(\alpha{+\beta}+\gamma \right)}{\left(\beta{+\gamma}\right)} & \beta & 0 & \displaystyle\frac{ra_{1}\left(\beta{+\gamma}\right)}{\mathrm{\mathcal{R}}_{0}\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ \alpha & -\left(\beta{+\gamma}\right)& 0 & 0 & 0\\ \displaystyle\frac{\gamma \left(\mathrm{\mathcal{R}}_{0}-1\right)\left(\alpha{+\beta}+\gamma \right)}{\left(\beta{+\gamma}\right)} & 0 & -A_{1} & \displaystyle\frac{ra_{1}\left(\beta{+\gamma}\right)}{\mathrm{\mathcal{R}}_{0}\left(\gamma{+}r\right)\left(\alpha{+\beta}+\gamma \right)} & 0\\ 0 & 0 & a_{2} & -A_{2} & 0\\ 0 & 0 & 0 & a_{3} & -\left(\gamma{+}b_{3}\right) \end{array}\right), </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (10) | | style="width: 5px;text-align: right;white-space: nowrap;" | (10) | ||
| Line 228: | Line 242: | ||
===3.2 The global asymptotic equilibrium stability=== | ===3.2 The global asymptotic equilibrium stability=== | ||
| − | ''' | + | '''Theorem 3.2.1'''. The disease-free equilibrium of the plant disease model is globally asymptotically stable in the suitable range if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and unstable if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math> . |
| − | '''Proof''' We are applying the Lyapunov function [33], which is defined by | + | '''Proof'''. We are applying the Lyapunov function [33], which is defined by |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 261: | Line 275: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \frac{1}{\ell _{1}}{}^{ABC}D_{*}^{\upsilon }S_{1}+\frac{1}{\ell _{2}}{}^{ABC}D_{*}^{\upsilon }P_{1}+\frac{1}{\ell _{3}}{}^{ABC}D_{*}^{\upsilon }E_{1}+\frac{1}{\ell _{4}}{}^{ABC}D_{*}^{\upsilon }I_{1}+\frac{1}{\ell _{5}}{}^{ABC}D_{*}^{\upsilon }R_{1}. </math> | + | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \frac{1}{\ell _{1}}{}^{ABC}D_{*}^{\upsilon }S_{1}+\frac{1}{\ell _{2}}{}^{ABC}D_{*}^{\upsilon }P_{1}+\frac{1}{\ell _{3}}{}^{ABC}D_{*}^{\upsilon }E_{1}</math> <math>+\frac{1}{\ell _{4}}{}^{ABC}D_{*}^{\upsilon }I_{1}+\frac{1}{\ell _{5}}{}^{ABC}D_{*}^{\upsilon }R_{1}. </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (14) | | style="width: 5px;text-align: right;white-space: nowrap;" | (14) | ||
| Line 271: | Line 285: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \frac{1}{\ell _{1}}\left[r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}-\left(\alpha{+\gamma}\right)S_{1}\right]+\frac{1}{\ell _{2}}\left[\alpha S_{1}-\left(\gamma{-\beta}\right)P_{1}\right] | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = \frac{1}{\ell _{1}}\left[r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}-\left(\alpha{+\gamma}\right)S_{1}\right]\\ |
| − | + | & \quad +\frac{1}{\ell _{2}}\left[\alpha S_{1}-\left(\gamma{-\beta}\right)P_{1}\right]\\ | |
| − | + | & \quad +\frac{1}{\ell_{3}}\left[\frac{a_{1}}{M}S_{1}I_{1}-\left(\gamma{+}a_{2}+b_{1}\right)E_{1}\right]\\ | |
| + | & \quad +\frac{1}{\ell _{4}}\left[a_{2}E_{1}-\left(\gamma{+}a_{3}+b_{2}+\eta \right)I_{1}\right]\\ | ||
| + | & \quad +\frac{1}{\ell _{5}}\left[a_{3}I_{1}-\left(\gamma{+}b_{3}\right)R_{1}\right], \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (15) | | style="width: 5px;text-align: right;white-space: nowrap;" | (15) | ||
| Line 283: | Line 299: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \frac{1}{\ell _{1}}\left[r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}-\ell _{1}S_{1}\right]+\frac{1}{\ell _{2}}\left[\alpha S_{1}-\ell _{2}P_{1}\right]+\frac{1}{\ell _{3}}\left[\frac{a_{1}}{M}S_{1}I_{1}-\ell _{3}E_{1}\right]+\frac{1}{\ell _{4}}\left[a_{2}E_{1}-\ell _{4}I_{1}\right]+\frac{1}{\ell _{5}}\left[a_{3}I_{1}-\ell _{5}R_{1}\right] | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = \, \frac{1}{\ell _{1}}\left[r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}-\ell _{1}S_{1}\right]+\frac{1}{\ell _{2}}\left[\alpha S_{1}-\ell _{2}P_{1}\right]+\frac{1}{\ell _{3}}\left[\frac{a_{1}}{M}S_{1}I_{1}-\ell _{3}E_{1}\right]\\ |
| − | + | & \quad + \frac{1}{\ell _{4}}\left[a_{2}E_{1}-\ell _{4}I_{1}\right]+\frac{1}{\ell _{5}}\left[a_{3}I_{1}-\ell _{5}R_{1}\right]\\ | |
| − | + | & = \, \left\{\frac{1}{\ell _{1}}\left(r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}\right)+\frac{\alpha }{\ell _{2}}S_{1}+\frac{1}{\ell _{3}}\frac{a_{1}}{M}S_{1}I_{1}+\frac{a_{2}}{\ell _{4}}E_{1}+\frac{a_{3}}{\ell _{5}}I_{1}\right\}\\ | |
| + | &\quad - \left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right)\\ | ||
| + | & =\, \Bigg[\left\{\frac{1}{\ell _{1}}\left(r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}\right)+\frac{\alpha }{\ell _{2}}S_{1}+\frac{1}{\ell _{3}}\frac{a_{1}}{M}S_{1}I_{1}+\frac{a_{2}}{\ell _{4}}E_{1}+\frac{a_{3}}{\ell _{5}}I_{1}\right\} \\ | ||
| + | & \quad\frac{1}{\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right)}-1\Bigg]\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right). \end{align}</math> | ||
| + | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (16) | | style="width: 5px;text-align: right;white-space: nowrap;" | (16) | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| Line 299: | Line 316: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \ | + | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = \Bigg[\left\{\frac{1}{\ell _{1}}\left(\frac{r(M-N)}{S_{1}}-\frac{a_{1}}{M}I_{1}+\beta \frac{P_{1}}{S_{1}}\right)+\frac{\alpha }{\ell _{2}}+\frac{1}{\ell _{3}}\frac{a_{1}}{M}I_{1}+\frac{a_{2}}{\ell _{4}}\frac{E_{1}}{S_{1}}+\frac{a_{3}}{\ell _{5}}\frac{I_{1}}{S_{1}}\right\}</math> |
| + | |- | ||
| + | | style="text-align: center;" |<math>\frac{1}{\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right)}-1\Bigg]\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right). </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (17) | | style="width: 5px;text-align: right;white-space: nowrap;" | (17) | ||
|} | |} | ||
| − | Since S is major than P, E, I and R categories, so | + | Since <math>S</math> is major than <math>P,\, E,\, I</math> and <math>R</math> categories, so |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 311: | Line 330: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} \leq \left[\frac{ra_{1}a_{2}(\beta{+\gamma})}{\left(a_{2}+b_{1}+\gamma \right)\left(\gamma{+}a_{3}+b_{2}+\eta \right)(\gamma{+}r)\left(\alpha{+\beta}+\gamma \right)}-1\right]\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right) | + | | style="text-align: center;" | <math>\begin{align}^{ABC}D_{*}^{\upsilon }\mathcal{L} & \leq \left[\frac{ra_{1}a_{2}(\beta{+\gamma})}{\left(a_{2}+b_{1}+\gamma \right)\left(\gamma{+}a_{3}+b_{2}+\eta \right)(\gamma{+}r)\left(\alpha{+\beta}+\gamma \right)}-1\right]\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right)\\ |
| − | + | &\leq \left[\mathrm{\mathcal{R}}_{0}-1\right]\left(S_{1}+P_{1}+E_{1}+I_{1}+R_{1}\right)\leq{0.} \end{align}</math> | |
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (18) | | style="width: 5px;text-align: right;white-space: nowrap;" | (18) | ||
|} | |} | ||
| − | Since <math display="inline">S_{1}+P_{1}+E_{1}+I_{1}+R_{1}>0,\forall t</math>. According to the proposed model, plant disease model would therefore be eliminated if and only if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math>. In general, because all parameters are positive in the plant disease model, Lyapunov function <math display="inline">\left(^{ABC}D_{*}^{\upsilon }\mathcal{L}\right)</math> therefore decreases if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and increases if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, eventually <math display="inline">\mathcal{L}=0</math> if <math display="inline">S_{1}=P_{1}=E_{1}=I_{1}=R_{1}=0</math>. <math display="inline">\mathcal{L}</math> is therefore the function of Lyapunov within the practicable biological interval and the greater compact invariant set in <math display="inline">\left\{S_{1},P_{1},E_{1},I_{1},R_{1}\in \Theta :{}^{ABC}D_{*}^{\upsilon }\mathcal{L}\leq{0}\right\}</math> is the point <math display="inline">\mathcal{E_{1}}_{0}</math>. Every solution of the plant disease model proposed in this study with an initial term in <math display="inline">\Theta </math> tends to <math display="inline">\mathcal{E_{1}}_{0}</math> when <math display="inline">t\rightarrow \infty </math> if and only if <math display="inline">\mathrm{\mathcal{R}}_{0}\leq{1}</math> through the well-known Lasalles invariance principle [ | + | Since <math display="inline">S_{1}+P_{1}+E_{1}+I_{1}+R_{1}>0,\forall t</math>. According to the proposed model, plant disease model would therefore be eliminated if and only if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math>. In general, because all parameters are positive in the plant disease model, Lyapunov function <math display="inline">\left(^{ABC}D_{*}^{\upsilon }\mathcal{L}\right)</math> therefore decreases if <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and increases if <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, eventually <math display="inline">\mathcal{L}=0</math> if <math display="inline">S_{1}=P_{1}=E_{1}=I_{1}=R_{1}=0</math>. <math display="inline">\mathcal{L}</math> is therefore the function of Lyapunov within the practicable biological interval and the greater compact invariant set in <math display="inline">\left\{S_{1},P_{1},E_{1},I_{1},R_{1}\in \Theta :{}^{ABC}D_{*}^{\upsilon }\mathcal{L}\leq{0}\right\}</math> is the point <math display="inline">\mathcal{E_{1}}_{0}</math>. Every solution of the plant disease model proposed in this study with an initial term in <math display="inline">\Theta </math> tends to <math display="inline">\mathcal{E_{1}}_{0}</math> when <math display="inline">t\rightarrow \infty </math> if and only if <math display="inline">\mathrm{\mathcal{R}}_{0}\leq{1}</math> through the well-known Lasalles invariance principle [38]. In conclusion, the plant disease model's disease-free equilibrium <math display="inline">\mathcal{E_{1}}_{0}</math> presented here is globally asymptotically stable. |
| − | ''' | + | '''Theorem 3.2.2'''. The endemic equilibrium point <math display="inline">\mathcal{E_{1}}^{*}</math> of the plant disease system is globally asymptotically stable if <math>\mathrm{\mathcal{R}}_{0}\leq{1}</math>. |
| − | '''Proof''' We use the Lyapunov function [ | + | '''Proof'''. We use the Lyapunov function [38] to prove this |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 329: | Line 347: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\mathcal{L}\left(S_{1}^{*},P_{1}^{*},E_{1}^{*},I_{1}^{*},R_{1}^{*}\right) | + | | style="text-align: center;" | <math>\begin{align}\mathcal{L}\left(S_{1}^{*},P_{1}^{*},E_{1}^{*},I_{1}^{*},R_{1}^{*}\right) &= \, \left(S_{1}-S_{1}^{*}-S_{1}^{*}\log \frac{S_{1}^{*}}{S_{1}}\right)+\left(P_{1}-P_{1}^{*}-P_{1}^{*}\log \frac{P_{1}^{*}}{P_{1}}\right)+\left(E_{1}-E_{1}^{*}-E_{1}^{*}\log \frac{E_{1}^{*}}{E_{1}}\right)\\ |
| − | + | &\quad + \left(I_{1}-I_{1}^{*}-I_{1}^{*}\log \frac{I_{1}^{*}}{I_{1}}\right)+\left(R_{1}-R_{1}^{*}-R_{1}^{*}\log \frac{R_{1}^{*}}{R_{1}}\right). \end{align}</math> | |
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (19) | | style="width: 5px;text-align: right;white-space: nowrap;" | (19) | ||
| Line 343: | Line 360: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L}=\left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right){}^{ABC}D_{*}^{\upsilon }S_{1}+\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right){}^{ABC}D_{*}^{\upsilon }P_{1}+\left(\frac{E_{1}-E_{1}^{*}}{E_{1}}\right){}^{ABC}D_{*}^{\upsilon }E_{1}+\left(\frac{I_{1}-I_{1}^{*}}{I_{1}}\right){}^{ABC}D_{*}^{\upsilon }I_{1}+\left(\frac{R_{1}-R_{1}^{*}}{R_{1}}\right){}^{ABC}D_{*}^{\upsilon }R_{1}, </math> | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = \left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right){}^{ABC}D_{*}^{\upsilon }S_{1}+\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right){}^{ABC}D_{*}^{\upsilon }P_{1}+\left(\frac{E_{1}-E_{1}^{*}}{E_{1}}\right){}^{ABC}D_{*}^{\upsilon }E_{1}\\ |
| + | &\quad + \left(\frac{I_{1}-I_{1}^{*}}{I_{1}}\right){}^{ABC}D_{*}^{\upsilon }I_{1}+\left(\frac{R_{1}-R_{1}^{*}}{R_{1}}\right){}^{ABC}D_{*}^{\upsilon }R_{1}, \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (20) | | style="width: 5px;text-align: right;white-space: nowrap;" | (20) | ||
| Line 355: | Line 373: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = \left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)\left(r(M-N)-\frac{a_{1}}{M}S_{1}I_{1}+\beta P_{1}-\ell _{1}S_{1}\right)\\ |
| − | + | &\quad +\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right)\left(\alpha S_{1}-\ell _{2}P_{1}\right) +\left(\frac{E_{1}-E_{1}^{*}}{E_{1}}\right)\left(\frac{a_{1}}{M}S_{1}I_{1}-\ell _{3}E_{1}\right)\\ | |
| − | + | &\quad +\left(\frac{I_{1}-I_{1}^{*}}{I_{1}}\right)\left(a_{2}E_{1}-\ell _{4}I_{1}\right)+\left(\frac{R_{1}-R_{1}^{*}}{R_{1}}\right)\left(a_{3}I_{1}-\ell _{5}R_{1}\right).\end{align} </math> | |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (21) | | style="width: 5px;text-align: right;white-space: nowrap;" | (21) | ||
| Line 369: | Line 387: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = \left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)\left[r(M-N)-\frac{a_{1}}{M}\left(S_{1}-S_{1}^{*}\right)\left(I_{1}-I_{1}^{*}\right)+\beta \left(P_{1}-P_{1}^{*}\right)-\ell _{1}\left(S_{1}-S_{1}^{*}\right)\right]\\ |
| − | + | &\quad +\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right)\left[\alpha \left(S_{1}-S_{1}^{*}\right)-\ell _{2}\left(P_{1}-P_{1}^{*}\right)\right]\\ | |
| − | + | &\quad +\left(\frac{E_{1}-E_{1}^{*}}{E_{1}}\right)\left[\frac{a_{1}}{M}\left(S_{1}-S_{1}^{*}\right)\left(I_{1}-I_{1}^{*}\right)-\ell _{3}\left(E_{1}-E_{1}^{*}\right)\right]\\ | |
| + | &\quad +\left(\frac{I_{1}-I_{1}^{*}}{I_{1}}\right)\left[a_{2}\left(E_{1}-E_{1}^{*}\right)-\ell _{4}\left(I_{1}-I_{1}^{*}\right)\right]\\ | ||
| + | &\quad +\left(\frac{R_{1}-R_{1}^{*}}{R_{1}}\right)\left[a_{3}\left(I_{1}-I_{1}^{*}\right)-\ell _{5}\left(R_{1}-R_{1}^{*}\right)\right].\qquad \end{align} </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (22) | | style="width: 5px;text-align: right;white-space: nowrap;" | (22) | ||
| Line 383: | Line 403: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = r(M-N)\left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)-\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)\left(\frac{a_{1}}{M}\left(I_{1}-I_{1}^{*}\right)+\ell _{1}\right)+\beta \left(P_{1}-P_{1}^{*}\right)\left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)+\alpha \left(S_{1}-S_{1}^{*}\right)\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right) | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = r(M-N)\left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)-\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)\left(\frac{a_{1}}{M}\left(I_{1}-I_{1}^{*}\right)+\ell _{1}\right)\\ |
| − | + | &\quad +\beta \left(P_{1}-P_{1}^{*}\right)\left(\frac{S_{1}-S_{1}^{*}}{S_{1}}\right)+\alpha \left(S_{1}-S_{1}^{*}\right)\left(\frac{P_{1}-P_{1}^{*}}{P_{1}}\right)\\ | |
| − | + | &\quad -\ell _{2}\left(\frac{\left(P_{1}-P_{1}^{*}\right)^{2}}{P_{1}}\right)+\frac{a_{1}}{M}\left(S_{1}-S_{1}^{*}\right)\left(I_{1}-I_{1}^{*}\right)\left(\frac{E_{1}-E_{1}^{*}}{E_{1}}\right)\\ | |
| + | &\quad -\ell_{3}\left(\frac{\left(E_{1}-E_{1}^{*}\right)^{2}}{E_{1}}\right)+a_{2}\left(E_{1}-E_{1}^{*}\right) \left(\frac{I_{1}-I_{1}^{*}}{I_{1}}\right)\\ | ||
| + | &\quad -\ell _{4}\left(\frac{\left(I_{1}-I_{1}^{*}\right)^{2}}{I_{1}}\right) + a_{3}\left(I_{1}-I_{1}^{*}\right)\left(\frac{R_{1}-R_{1}^{*}}{R_{1}}\right)\\ | ||
| + | &\quad -\ell _{5}\left(\frac{\left(R_{1}-R_{1}^{*}\right)^{2}}{R_{1}}\right),\end{align} </math> | ||
| + | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (23) | | style="width: 5px;text-align: right;white-space: nowrap;" | (23) | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| Line 397: | Line 418: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>^{ABC}D_{*}^{\upsilon }\mathcal{L} = r(M-N)-r(M-N)\frac{S_{1}^{*}}{S_{1}}-\frac{a_{1}}{M}I_{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\frac{a_{1}}{M}I_{1}^{*}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)-\ell _{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}-\beta P_{1}^{*}-\beta P_{1}\frac{S_{1}^{*}}{S_{1}}+\beta P_{1}^{*}\frac{S_{1}^{*}}{S_{1}}+\alpha S_{1}-\alpha S_{1}^{*} | + | | style="text-align: center;" | <math>\begin{align} ^{ABC}D_{*}^{\upsilon }\mathcal{L} & = r(M-N)-r(M-N)\frac{S_{1}^{*}}{S_{1}}-\frac{a_{1}}{M}I_{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\frac{a_{1}}{M}I_{1}^{*}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)\\ |
| − | + | &\quad -\ell _{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}-\beta P_{1}^{*}-\beta P_{1}\frac{S_{1}^{*}}{S_{1}}+\beta P_{1}^{*}\frac{S_{1}^{*}}{S_{1}} +\alpha S_{1}-\alpha S_{1}^{*}-\alpha S_{1}\frac{P_{1}^{*}}{P_{1}}\\ | |
| − | + | &\quad +\alpha S_{1}^{*}\frac{P_{1}^{*}}{P_{1}}-\ell _{2}\left(\frac{\left(P_{1}-P_{1}^{*}\right)^{2}}{P_{1}}\right)+\frac{a_{1}}{M}S_{1}I_{1}-\frac{a_{1}}{M}S_{1}I_{1}^{*}-\frac{a_{1}}{M}S_{1}I_{1}\frac{E_{1}^{*}}{E_{1}}\\ | |
| + | &\quad +\frac{a_{1}}{M}S_{1}I_{1}^{*}\frac{E_{1}^{*}}{E_{1}}-\frac{a_{1}}{M}S_{1}^{*}I_{1}+\frac{a_{1}}{M}S_{1}^{*}I_{1}^{*}+\frac{a_{1}}{M}S_{1}^{*}I_{1}\frac{E_{1}^{*}}{E_{1}}-\frac{a_{1}}{M}S_{1}^{*}I_{1}^{*}\frac{E_{1}^{*}}{E_{1}}\\ | ||
| + | &\quad -\ell _{3}\left(\frac{\left(E_{1}-E_{1}^{*}\right)^{2}}{E_{1}}\right)+a_{2}E_{1}-a_{2}E_{1}^{*}-a_{2}E_{1}\frac{I_{1}^{*}}{I_{1}}+a_{2}E_{1}^{*}\frac{I_{1}^{*}}{I_{1}}-\ell _{4}\left(\frac{\left(I_{1}-I_{1}^{*}\right)^{2}}{I_{1}}\right)\\ | ||
| + | &\quad +a_{3}I_{1}-a_{3}I_{1}^{*}-a_{3}I_{1}\frac{R_{1}^{*}}{R_{1}}+a_{3}I_{1}^{*}\frac{R_{1}^{*}}{R_{1}}-\ell _{5}\left(\frac{\left(R_{1}-R_{1}^{*}\right)^{2}}{R_{1}}\right).\end{align} </math> | ||
| + | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (24) | | style="width: 5px;text-align: right;white-space: nowrap;" | (24) | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| Line 425: | Line 447: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\mathcal{L}_{1} = r(M-N)+\frac{a_{1}}{M}I_{1}^{*}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}+\beta P_{1}^{*}\frac{S_{1}^{*}}{S_{1}}+\alpha S_{1}+\alpha S_{1}^{*}\frac{P_{1}^{*}}{P_{1}}+\frac{a_{1}}{M}S_{1}I_{1}+\frac{a_{1}}{M}S_{1}I_{1}^{*}\frac{E_{1}^{*}}{E_{1}} | + | | style="text-align: center;" | <math>\begin{align}\mathcal{L}_{1} & = r(M-N)+\frac{a_{1}}{M}I_{1}^{*}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}+\beta P_{1}^{*}\frac{S_{1}^{*}}{S_{1}}+\alpha S_{1}\\ |
| − | + | &\quad +\alpha S_{1}^{*}\frac{P_{1}^{*}}{P_{1}}+\frac{a_{1}}{M}S_{1}I_{1}+\frac{a_{1}}{M}S_{1}I_{1}^{*}\frac{E_{1}^{*}}{E_{1}} +\frac{a_{1}}{M}S_{1}^{*}I_{1}^{*}\\ | |
| − | + | &\quad +\frac{a_{1}}{M}S_{1}^{*}I_{1}\frac{E_{1}^{*}}{E_{1}}+a_{2}E_{1}+a_{2}E_{1}^{*}\frac{I_{1}^{*}}{I_{1}}+a_{3}I_{1}+a_{3}I_{1}^{*}\frac{R_{1}^{*}}{R_{1}}, \end{align}</math> | |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (26) | | style="width: 5px;text-align: right;white-space: nowrap;" | (26) | ||
| Line 439: | Line 461: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\mathcal{L}_{2} = r(M-N)\frac{S_{1}^{*}}{S_{1}}+\frac{a_{1}}{M}I_{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\ell _{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}^{*}+\beta P_{1}\frac{S_{1}^{*}}{S_{1}}+\alpha S_{1}^{*}+\alpha S_{1}\frac{P_{1}^{*}}{P_{1}}+\ell _{2}\left(\frac{\left(P_{1}-P_{1}^{*}\right)^{2}}{P_{1}}\right)+\frac{a_{1}}{M}S_{1}I_{1}^{*}+\frac{a_{1}}{M}S_{1}I_{1}\frac{E_{1}^{*}}{E_{1}} | + | | style="text-align: center;" | <math>\begin{align} \mathcal{L}_{2} & = r(M-N)\frac{S_{1}^{*}}{S_{1}}+\frac{a_{1}}{M}I_{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\ell _{1}\left(\frac{\left(S_{1}-S_{1}^{*}\right)^{2}}{S_{1}}\right)+\beta P_{1}^{*}+\beta P_{1}\frac{S_{1}^{*}}{S_{1}}\\ |
| − | + | &\quad +\alpha S_{1}^{*}+\alpha S_{1}\frac{P_{1}^{*}}{P_{1}}+\ell _{2}\left(\frac{\left(P_{1}-P_{1}^{*}\right)^{2}}{P_{1}}\right)+\frac{a_{1}}{M}S_{1}I_{1}^{*}+\frac{a_{1}}{M}S_{1}I_{1}\frac{E_{1}^{*}}{E_{1}}\\ | |
| − | + | &\quad +\frac{a_{1}}{M}S_{1}^{*}I_{1}+\frac{a_{1}}{M}S_{1}^{*}I_{1}^{*}\frac{E_{1}^{*}}{E_{1}}+\ell _{3}\left(\frac{\left(E_{1}-E_{1}^{*}\right)^{2}}{E_{1}}\right)+a_{2}E_{1}^{*}+a_{2}E_{1}\frac{I_{1}^{*}}{I_{1}}\\ | |
| + | &\quad +\ell _{4}\left(\frac{\left(I_{1}-I_{1}^{*}\right)^{2}}{I_{1}}\right)+a_{3}I_{1}^{*}+a_{3}I_{1}\frac{R_{1}^{*}}{R_{1}}+\ell _{5}\left(\frac{\left(R_{1}-R_{1}^{*}\right)^{2}}{R_{1}}\right),\end{align} </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (27) | | style="width: 5px;text-align: right;white-space: nowrap;" | (27) | ||
| Line 450: | Line 473: | ||
We can now conclude that the largest compact invariant set for the plant diseases model in <math display="inline">\left\{S_{1}^{*},P_{1}^{*},E_{1}^{*},I_{1}^{*},R_{1}^{*}\in \Theta :{}^{ABC}D_{*}^{\upsilon }\mathcal{L}=0\right\}</math> is the point <math display="inline">\mathcal{E}^{*}</math> the endemic equilibrium of the plant diseases model. | We can now conclude that the largest compact invariant set for the plant diseases model in <math display="inline">\left\{S_{1}^{*},P_{1}^{*},E_{1}^{*},I_{1}^{*},R_{1}^{*}\in \Theta :{}^{ABC}D_{*}^{\upsilon }\mathcal{L}=0\right\}</math> is the point <math display="inline">\mathcal{E}^{*}</math> the endemic equilibrium of the plant diseases model. | ||
| − | ===3.3 <span id='lb-3.3'></span>Sensibility analysis of reproduction number ( | + | ===3.3 <span id='lb-3.3'></span>Sensibility analysis of reproduction number (<math>\mathrm{\mathcal{R}}_{0}</math> ) without control=== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Since the parameters of the epizootic system are either predestined or equipped, this raises some doubt as to their values used to draw a conclusion about the underlying epidemic. Therefore, it's critical to identify the specific effects of each element on the dynamics of the pestilence and group the variables that have the most impact to limit or spread the outbreak. Within this section, a sensitivity analysis is conducted for the disease parameters identified with the suggested SPEIR model via the sensitivity indicator. Quantifying the most sensitive aspects of the basal reproductive number <math display="inline">\mathrm{\mathcal{R}}_{0}</math> can be done with the help of the Sensitivity Indicator strategy. The following equations give the standardized sensitivity indicator <math display="inline">\Psi _{*}^{\mathrm{\mathcal{R}}_{0}}</math> of <math display="inline">\mathrm{\mathcal{R}}_{0}</math> for all the parameters <math display="inline">\left(r,a_{1},a_{2},a_{3},b_{1},b_{2},\alpha ,\beta ,\gamma ,\eta \right)</math> used within the SPEIR model in [[#table-1|Table 1]] where <math display="inline">\Psi _{*}^{\mathrm{\mathcal{R}}_{0}}=\frac{\partial \mathrm{\mathcal{R}}_{0}}{\partial }\times \frac{*}{\left|\mathrm{\mathcal{R}}_{0}\right|}</math> | ||
<span id="eq-28"></span> | <span id="eq-28"></span> | ||
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
|- | |- | ||
| | | | ||
| − | { | + | {| style="text-align: center; margin:auto;width: 100%;" |
| − | + | ||
| − | | style="text-align: center | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
|- | |- | ||
| − | | style="text-align: center;" | <math> \Psi _{\gamma }^{\mathrm{\mathcal{R}}_{0}}=\frac{a_{1}a_{2}r\left(-\alpha \beta{-}(\beta{+\gamma})^{2}+\alpha r\right)}{A_{1}A_{2}(\gamma{+}r)^{2}(\alpha{+\beta}+\gamma )^{2}}-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}^{2}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}^{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=-0.312737<0, </math> | + | | style="text-align: center;" | <math>\begin{align} \Psi _{r}^{\mathrm{\mathcal{R}}_{0}} & =\frac{a_{1}a_{2}\gamma (\beta{+\gamma})}{A_{1}A_{2}(\gamma{+}r)^{2}(\alpha{+\beta}+\gamma )}=0.266667>0, \\ |
| + | \Psi _{a_{1}}^{\mathrm{\mathcal{R}}_{0}} & =\frac{a_{2}r(\beta{+\gamma})}{A_{1}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=1>0,\\ | ||
| + | \Psi _{a_{2}}^{\mathrm{\mathcal{R}}_{0}} & =\frac{a_{1}r(\beta{+\gamma})\left(b_{1}+\gamma \right)}{A_{1}^{2}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=0.0229885>0,\\ | ||
| + | \Psi _{a_{3}}^{\mathrm{\mathcal{R}}_{0}} & =-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}^{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=-0.300752<0,\\ | ||
| + | \Psi _{b_{1}}^{\mathrm{\mathcal{R}}_{0}} & =-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}^{2}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=0, \\ | ||
| + | \Psi _{b_{2}}^{\mathrm{\mathcal{R}}_{0}} & =-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}^{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=0,\\ | ||
| + | \Psi _{\alpha }^{\mathrm{\mathcal{R}}_{0}}& =-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )^{2}}=-0.0909091<0, \\ | ||
| + | \Psi _{\beta }^{\mathrm{\mathcal{R}}_{0}}& =\frac{\alpha a_{1}a_{2}r}{A_{1}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )^{2}}=0.0839161>0, \\ | ||
| + | \Psi _{\eta }^{\mathrm{\mathcal{R}}_{0}}& =-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}^{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=-0.669173<0,\\ | ||
| + | \Psi _{\gamma }^{\mathrm{\mathcal{R}}_{0}} & =\frac{a_{1}a_{2}r\left(-\alpha \beta{-}(\beta{+\gamma})^{2}+\alpha r\right)}{A_{1}A_{2}(\gamma{+}r)^{2}(\alpha{+\beta}+\gamma )^{2}}-\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}^{2}A_{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}\\ | ||
| + | &\quad -\frac{a_{1}a_{2}r(\beta{+\gamma})}{A_{1}A_{2}^{2}(\gamma{+}r)(\alpha{+\beta}+\gamma )}=-0.312737<0,\end{align}</math> | ||
|} | |} | ||
| + | | style="width: 5px;text-align: right;white-space: nowrap;" |(28) | ||
|} | |} | ||
| − | As shown in the previous calculations, some component of the sensitivity indicator are positive, like <math>r</math>, <math>a_{1}</math>, <math>a_{2}</math> and <math>\beta </math>, while others, like <math>a_{3}</math>, <math>\alpha </math>, <math>\gamma </math> and <math>\eta </math> are negative. Furthermore, the most important feature of these indicators is the functionality of the SPEIR model parameters. This means that getting a small amendment in one of the parameters will amendment the epidemic dynamics. The value <math>\Psi _{a_{3}}^{\mathrm{\mathcal{R}}_{0}}=-0.300752</math> displays that decreasing (increasing) <math>a_{3}</math> for example by 70% increases (decreases) the basic reproductive number <math>\mathrm{\mathcal{R}}_{0}</math> by about 70% A small change in a parameter can head to comparatively enormous quantitative changes, requiring these sensitive parameters to be understood. It can be shown from previous calculations that parameters <math>a_{1}</math> (disease progression diversion rate for Latent Compartmentenglish) and <math>\eta </math> (rate of roguingenglish), respectively, | + | As shown in the previous calculations, some component of the sensitivity indicator are positive, like <math>r</math>, <math>a_{1}</math>, <math>a_{2}</math> and <math>\beta </math>, while others, like <math>a_{3}</math>, <math>\alpha </math>, <math>\gamma </math> and <math>\eta </math> are negative. Furthermore, the most important feature of these indicators is the functionality of the SPEIR model parameters. This means that getting a small amendment in one of the parameters will amendment the epidemic dynamics. The value <math>\Psi _{a_{3}}^{\mathrm{\mathcal{R}}_{0}}=-0.300752</math> displays that decreasing (increasing) <math>a_{3}</math> for example by 70% increases (decreases) the basic reproductive number <math>\mathrm{\mathcal{R}}_{0}</math> by about 70% A small change in a parameter can head to comparatively enormous quantitative changes, requiring these sensitive parameters to be understood. It can be shown from previous calculations that parameters <math>a_{1}</math> (disease progression diversion rate for Latent Compartmentenglish) and <math>\eta </math> (rate of roguingenglish) are, respectively, the maximum and minimum sensitivity epidemical parameters <math>\mathrm{\mathcal{R}}_{0}</math>. |
| − | ==4 Achieve existence and uniqueness== | + | ==4. Achieve existence and uniqueness== |
In this section, we will prove that model 4 has a unique solution, that the kernel satisfies Lipschitz's condition and that the functions in this model is bounded. | In this section, we will prove that model 4 has a unique solution, that the kernel satisfies Lipschitz's condition and that the functions in this model is bounded. | ||
| − | Now, we will analyze the fractional model ([[#eq-1|1]]). Usage of an integral fractional operator on Eq. ([[#eq-1|1]]), we are gaining | + | Now, we will analyze the fractional model (Eq.[[#eq-1|(1)]]). Usage of an integral fractional operator on Eq. ([[#eq-1|1]]), we are gaining |
<span id="eq-29"></span> | <span id="eq-29"></span> | ||
| Line 592: | Line 543: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\begin{ | + | | style="text-align: center;" | <math>\begin{align} \left\Vert \varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right\Vert & = \left\Vert -\gamma \left(S_{1}-\tilde{S_{1}}\right)-\frac{a_{1}I_{1}}{M}\left(S_{1}-\tilde{S_{1}}\right)-\alpha \left(S_{1}-\tilde{S_{1}}\right)\right\Vert \\ & \leq \gamma \left\Vert S_{1}-\tilde{S_{1}}\right\Vert +\frac{a_{1}\phi }{M}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert +\alpha \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \\ & \leq \left\{\gamma +\frac{a_{1}\phi }{M}+\alpha \right\}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert , \end{align}</math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (31) | | style="width: 5px;text-align: right;white-space: nowrap;" | (31) | ||
| Line 636: | Line 587: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>X_{S_{1}}=\gamma +\frac{a_{1}\phi }{M}+\alpha ,\ | + | | style="text-align: center;" | <math>\begin{align} X_{S_{1}} & =\gamma +\frac{a_{1}\phi }{M}+\alpha ,\\ |
| + | X_{P_{1}}& =\beta{+\gamma},\\ | ||
| + | X_{E_{1}}& =\gamma{+}a_{2}+b_{1},\\ | ||
| + | X_{I_{1}}& =\gamma{+}a_{3}+b_{2}+\eta ,\\ | ||
| + | X_{R_{1}}& =\gamma{+}b_{3}, \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (36) | | style="width: 5px;text-align: right;white-space: nowrap;" | (36) | ||
| Line 643: | Line 598: | ||
and <math>\left\Vert P_{1}\right\Vert \le c_{3}</math>, <math>\left\Vert \tilde{P_{1}}\right\Vert \le c_{4}</math>, <math>\left\Vert E_{1}\right\Vert \le c_{5}</math>, <math>\left\Vert \tilde{E_{1}}\right\Vert \le c_{6}</math>, <math>\left\Vert I_{1}\right\Vert \le c_{7}</math>, <math>\left\Vert \tilde{I_{1}}\right\Vert \le c_{8}</math>, <math>\left\Vert R_{1}\right\Vert \le c_{9}</math>, <math>\left\Vert \tilde{R_{1}}\right\Vert \le c_{10}</math>. Hence, for the kernels <math>\varrho _{1}\left(t,S_{1}\right)</math>, <math>\varrho _{2}\left(t,P_{1}\right)</math>, <math>\varrho _{3}\left(t,E_{1}\right)</math>, <math>\varrho _{4}\left(t,I_{1}\right)</math> and <math>\varrho _{5}\left(t,R_{1}\right)</math>, the Lipschitz condition is justified. | and <math>\left\Vert P_{1}\right\Vert \le c_{3}</math>, <math>\left\Vert \tilde{P_{1}}\right\Vert \le c_{4}</math>, <math>\left\Vert E_{1}\right\Vert \le c_{5}</math>, <math>\left\Vert \tilde{E_{1}}\right\Vert \le c_{6}</math>, <math>\left\Vert I_{1}\right\Vert \le c_{7}</math>, <math>\left\Vert \tilde{I_{1}}\right\Vert \le c_{8}</math>, <math>\left\Vert R_{1}\right\Vert \le c_{9}</math>, <math>\left\Vert \tilde{R_{1}}\right\Vert \le c_{10}</math>. Hence, for the kernels <math>\varrho _{1}\left(t,S_{1}\right)</math>, <math>\varrho _{2}\left(t,P_{1}\right)</math>, <math>\varrho _{3}\left(t,E_{1}\right)</math>, <math>\varrho _{4}\left(t,I_{1}\right)</math> and <math>\varrho _{5}\left(t,R_{1}\right)</math>, the Lipschitz condition is justified. | ||
| − | ''' | + | '''Theorem 4.1'''. Presume that <math display="inline">S_{1}\left(t\right)</math> is obliged, then the operator <math display="inline">\xi \left\{S_{1}\left(t\right)\right\}</math> is supplied by |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 657: | Line 612: | ||
satisfies the Lipschitz condition. | satisfies the Lipschitz condition. | ||
| − | '''Proof''' Assume that <math>S_{1}\left(t\right)</math> and <math>\tilde{S_{1}}\left(t\right)</math> are bounded functions with <math display="inline">S_{1}\left(0\right)=\tilde{S_{1}}\left(0\right)</math>, then we have | + | '''Proof'''. Assume that <math>S_{1}\left(t\right)</math> and <math>\tilde{S_{1}}\left(t\right)</math> are bounded functions with <math display="inline">S_{1}\left(0\right)=\tilde{S_{1}}\left(0\right)</math>, then we have |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 664: | Line 619: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert \xi \left\{S_{1}\left(t\right)\right\}-\xi \left\{\tilde{S_{1}}\left(t\right)\right\}\right\Vert | + | | style="text-align: center;" | <math>\begin{align} \left\Vert \xi \left\{S_{1}\left(t\right)\right\}-\xi \left\{\tilde{S_{1}}\left(t\right)\right\}\right\Vert & = \left\Vert \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left(\varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right)+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left(\varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right)d\theta \right\Vert \\ |
| − | + | & \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)} \left\Vert \varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right\Vert +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \varrho _{1} \left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right\Vert d\theta\\ | |
| − | + | & \leq \left(\frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}-\frac{X_{S_{1}}t^{\upsilon }}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right)\left\Vert S_{1}-\tilde{S_{1}}\right\Vert .\end{align} </math> | |
| − | + | ||
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (38) | | style="width: 5px;text-align: right;white-space: nowrap;" | (38) | ||
| Line 675: | Line 628: | ||
This completes the proof. The same process can be applied to <math display="inline">P_{1}\left(t\right)</math>, <math display="inline">E_{1}\left(t\right)</math>, <math display="inline">I_{1}\left(t\right)</math> and <math display="inline">R_{1}\left(t\right)</math>. | This completes the proof. The same process can be applied to <math display="inline">P_{1}\left(t\right)</math>, <math display="inline">E_{1}\left(t\right)</math>, <math display="inline">I_{1}\left(t\right)</math> and <math display="inline">R_{1}\left(t\right)</math>. | ||
| − | ''' | + | '''Theorem 4.2'''. If <math display="inline">S_{1}\left(t\right)</math> is a bounded, then the operator |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 715: | Line 668: | ||
|} | |} | ||
| − | '''Proof<math>\;</math>(i)''' Suppose that <math>S_{1}\left(t\right)</math> is bounded function, then | + | '''Proof<math>\;</math>(i)'''. Suppose that <math>S_{1}\left(t\right)</math> is bounded function, then |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 722: | Line 675: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\begin{ | + | | style="text-align: center;" | <math>\begin{align} \left|\left\langle \xi _{1}\left(S_{1}\right)-\xi _{1}\left(\tilde{S_{1}}\right),S_{1}-\tilde{S_{1}}\right\rangle \right|& = \left|\left\langle -\gamma \left(S_{1}-\tilde{S_{1}}\right)-\frac{a_{1}I_{1}}{M}\left(S_{1}-\tilde{S_{1}}\right)-\alpha \left(S_{1}-\tilde{S_{1}}\right),S_{1}-\tilde{S_{1}}\right\rangle \right|\\ & \leq \left|\left\langle \gamma \left(S_{1}-\tilde{S_{1}}\right),S_{1}-\tilde{S_{1}}\right\rangle \right|+\left|\left\langle \frac{a_{1}I_{1}}{M}\left(S_{1}-\tilde{S_{1}}\right),S_{1}-\tilde{S_{1}}\right\rangle \right|+\left|\left\langle \alpha \left(S_{1}-\tilde{S_{1}}\right),S_{1}-\tilde{S_{1}}\right\rangle \right|\\ & \leq \gamma \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert S_{1}-\tilde{S_{1}}\right\Vert +\frac{a_{1}\phi }{M}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert S_{1}-\tilde{S_{1}}\right\Vert +\alpha \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \\ & \leq \left\{\gamma +\frac{a_{1}\phi }{M}+\alpha \right\}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert ^{2}\\ & \leq X_{S_{1}}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert ^{2}. \end{align}</math> |
|} | |} | ||
| + | | style="width: 5px;text-align: right;white-space: nowrap;" | (42) | ||
|} | |} | ||
| − | '''Proof<math>\;</math>(ii)''' Suppose that <math display="inline">0<\left\Vert D\right\Vert <\infty </math>, since <math>S_{1}\left(t\right)</math> is bounded function, so | + | '''Proof<math>\;</math>(ii)'''. Suppose that <math display="inline">0<\left\Vert D\right\Vert <\infty </math>, since <math>S_{1}\left(t\right)</math> is bounded function, so |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 733: | Line 687: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\begin{ | + | | style="text-align: center;" | <math>\begin{align} \left|\left\langle \xi _{1}\left(S_{1}\right)-\xi _{1}\left(\tilde{S_{1}}\right),D\right\rangle \right|& = \left|\left\langle -\gamma \left(S_{1}-\tilde{S_{1}}\right)-\frac{a_{1}I_{1}}{M}\left(S_{1}-\tilde{S_{1}}\right)-\alpha \left(S_{1}-\tilde{S_{1}}\right),D\right\rangle \right|\\ & \leq \left|\left\langle \gamma \left(S_{1}-\tilde{S_{1}}\right),D\right\rangle \right|+\left|\left\langle \frac{a_{1}I_{1}}{M}\left(S_{1}-\tilde{S_{1}}\right),D\right\rangle \right|+\left|\left\langle \alpha \left(S_{1}-\tilde{S_{1}}\right),D\right\rangle \right|\\ & \leq \gamma \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert D\right\Vert +\frac{a_{1}\phi }{M}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert D\right\Vert +\alpha \left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert D\right\Vert \\ & \leq \left\{\gamma +\frac{a_{1}\phi }{M}+\alpha \right\}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert D\right\Vert \\ & \leq X_{S_{1}}\left\Vert S_{1}-\tilde{S_{1}}\right\Vert \left\Vert D\right\Vert . \end{align}</math> |
|} | |} | ||
| + | | style="width: 5px;text-align: right;white-space: nowrap;" | (43) | ||
|} | |} | ||
This completes the proof. The same process can be applied to <math display="inline">P_{1}\left(t\right)</math>, <math display="inline">E_{1}\left(t\right)</math>, <math display="inline">I_{1}\left(t\right)</math> and <math display="inline">R_{1}\left(t\right)</math>. | This completes the proof. The same process can be applied to <math display="inline">P_{1}\left(t\right)</math>, <math display="inline">E_{1}\left(t\right)</math>, <math display="inline">I_{1}\left(t\right)</math> and <math display="inline">R_{1}\left(t\right)</math>. | ||
| − | An inquiry into the existence and uniqueness of Eq. ([[#eq-1|1]]) will be discussed in the following. From Eq. ([[#eq-29|29]]) we can write | + | An inquiry into the existence and uniqueness of Eq.([[#eq-1|1]]) will be discussed in the following. From Eq. ([[#eq-29|29]]) we can write |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 759: | Line 714: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\Upsilon _{n+1}\left(t\right)=\left(S_{1}\right)_{n+1}-\left(S_{1}\right)_{n}=\frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right]+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right]d\theta{.} </math> | + | | style="text-align: center;" | <math>\begin{align}\Upsilon _{n+1}\left(t\right) & =\left(S_{1}\right)_{n+1}-\left(S_{1}\right)_{n} =\frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right]\\ |
| + | & \quad +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right]d\theta{.}\end{align} </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (45) | | style="width: 5px;text-align: right;white-space: nowrap;" | (45) | ||
|} | |} | ||
| − | According to [34], it would be easy to write | + | According to Castillo-Chavez et al. [34], it would be easy to write |
<span id="eq-46"></span> | <span id="eq-46"></span> | ||
| Line 784: | Line 740: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert =\left\Vert \left(S_{1}\right)_{n+1}-\left(S_{1}\right)_{n}\right\Vert =\left\Vert \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right]+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right]d\theta \right\Vert . </math> | + | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert =\left\Vert \left(S_{1}\right)_{n+1}-\left(S_{1}\right)_{n}\right\Vert =\left\Vert \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right]\right\Vert.</math><math>\left\Vert.+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right]d\theta \right\Vert . </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (47) | | style="width: 5px;text-align: right;white-space: nowrap;" | (47) | ||
| Line 796: | Line 752: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\Vert \varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right\Vert +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right\Vert d\theta{.} </math> | + | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\Vert \varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n-1}\right)\right\Vert</math><math> +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n-1}\right)\right\Vert d\theta{.} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (48) | | style="width: 5px;text-align: right;white-space: nowrap;" | (48) | ||
| Line 809: | Line 765: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}\left\Vert \left(S_{1}\right)_{n}-\left(S_{1}\right)_{n-1}\right\Vert +\frac{\upsilon X_{S_{1}}}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \left(S_{1}\right)_{n}-\left(S_{1}\right)_{n-1}\right\Vert d\theta{.} </math> | + | | style="text-align: center;" | <math>\left\Vert \Upsilon _{n+1}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}\left\Vert \left(S_{1}\right)_{n}-\left(S_{1}\right)_{n-1}\right\Vert</math><math> +\frac{\upsilon X_{S_{1}}}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \left(S_{1}\right)_{n}-\left(S_{1}\right)_{n-1}\right\Vert d\theta{.} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (49) | | style="width: 5px;text-align: right;white-space: nowrap;" | (49) | ||
|} | |} | ||
| − | ''' | + | '''Theorem 4.3'''. If <math display="inline">t_{0}</math> fits the following condition |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 826: | Line 782: | ||
|} | |} | ||
| − | then, model ([[#eq-1|1]]) has a unique solution. | + | then, model (Eq.[[#eq-1|(1)]]) has a unique solution. |
| − | '''Proof''' Suppose that <math display="inline">S_{1}(t)</math> is bounded. As the Lipschitz condition is fulfilled by the kernel, therefore, utilizing the recursive process of Eq. ([[#eq-49|49]]), we acquire | + | '''Proof'''. Suppose that <math display="inline">S_{1}(t)</math> is bounded. As the Lipschitz condition is fulfilled by the kernel, therefore, utilizing the recursive process of Eq. ([[#eq-49|49]]), we acquire |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 839: | Line 795: | ||
|} | |} | ||
| − | Therefore, the <math display="inline">S_{1}(t)</math> function offered by Eq. ([[#eq-46|46]]) exists and is smooth as well. Currently, we wish to illustrate that the above-mentioned functions are actually a solution to englishmodel ([[#eq-1|1]]). Presume that | + | Therefore, the <math display="inline">S_{1}(t)</math> function offered by Eq. ([[#eq-46|46]]) exists and is smooth as well. Currently, we wish to illustrate that the above-mentioned functions are actually a solution to englishmodel (Eq.[[#eq-1|(1)]]). Presume that |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 858: | Line 814: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert \left(\overline{S_{1}}\right)_{n}\right\Vert = \left\Vert \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)\right]+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)\right]d\theta \right\Vert | + | | style="text-align: center;" | <math>\begin{align}\left\Vert \left(\overline{S_{1}}\right)_{n}\right\Vert &= \left\Vert \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)\right]+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)\right]d\theta \right\Vert \\ |
| − | + | & \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}\Bigl\Vert \varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\left(S_{1}\right)_{n}\right)\Bigr\Vert +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\Bigl\Vert \varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\left(S_{1}\right)_{n}\right)\Bigr\Vert d\theta \\ | |
| − | + | & \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}\left\Vert S_{1}-\left(S_{1}\right)_{n}\right\Vert +\frac{\upsilon X_{S_{1}}t^{\upsilon }}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left\Vert S_{1}-\left(S_{1}\right)_{n}\right\Vert . \end{align}</math> | |
| + | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (52) | | style="width: 5px;text-align: right;white-space: nowrap;" | (52) | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| Line 892: | Line 846: | ||
|} | |} | ||
| − | Using the limit in Eq. ([[#eq-54|54]]) as <math display="inline">n</math> gets closer to <math display="inline">\infty </math> , we arrive at <math display="inline">\left\Vert \left(\overline{S_{1}}\right)_{n}\right\Vert \rightarrow{0}</math>. Thus, the existence is demonstrated. It is still necessary to demonstrate the uniqueness of the model english([[#eq-1|1]]). Assume that <math display="inline">\tilde{S_{1}}\left(t\right)</math> is another solution of model ([[#eq-1|1]]), then | + | Using the limit in Eq. ([[#eq-54|54]]) as <math display="inline">n</math> gets closer to <math display="inline">\infty </math> , we arrive at <math display="inline">\left\Vert \left(\overline{S_{1}}\right)_{n}\right\Vert \rightarrow{0}</math>. Thus, the existence is demonstrated. It is still necessary to demonstrate the uniqueness of the model english (Eq.[[#eq-1|(1)]]). Assume that <math display="inline">\tilde{S_{1}}\left(t\right)</math> is another solution of model (Eq.[[#eq-1|(1)]]), then |
<span id="eq-55"></span> | <span id="eq-55"></span> | ||
| Line 900: | Line 854: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right) = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right]+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right]d\theta{.} </math> | + | | style="text-align: center;" | <math>S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right) = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left[\varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right]</math><math>+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left[\varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right]d\theta{.} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (55) | | style="width: 5px;text-align: right;white-space: nowrap;" | (55) | ||
| Line 912: | Line 866: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\Vert \varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right\Vert +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right\Vert d\theta{.} </math> | + | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\Vert \varrho _{1}\left(t,S_{1}\right)-\varrho _{1}\left(t,\tilde{S_{1}}\right)\right\Vert </math><math>+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert \varrho _{1}\left(\theta ,S_{1}\right)-\varrho _{1}\left(\theta ,\tilde{S_{1}}\right)\right\Vert d\theta{.} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (56) | | style="width: 5px;text-align: right;white-space: nowrap;" | (56) | ||
| Line 924: | Line 878: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert +\frac{\upsilon X_{S_{1}}}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert d\theta{.} </math> | + | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert \leq \frac{1-\upsilon }{\chi \left(\upsilon \right)}X_{S_{1}}\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert </math><math>+\frac{\upsilon X_{S_{1}}}{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t-\theta \right)^{\upsilon{-1}}\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert d\theta{.} </math> |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (57) | | style="width: 5px;text-align: right;white-space: nowrap;" | (57) | ||
| Line 960: | Line 914: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert = 0, | + | | style="text-align: center;" | <math>\left\Vert S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right)\right\Vert = 0,\qquad S_{1}\left(t\right)-\tilde{S_{1}}\left(t\right) = 0,\qquad S_{1}\left(t\right) = \tilde{S_{1}}\left(t\right). </math> |
| − | + | |} | |
| − | + | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (60) | | style="width: 5px;text-align: right;white-space: nowrap;" | (60) | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | Thus, the uniqueness is verified. We can prove the uniqueness of the rest of equations in model ([[#eq-1|1]]) by using the same method. Therefore model ([[#eq-1|1]]) | + | Thus, the uniqueness is verified. We can prove the uniqueness of the rest of equations in model (Eq.[[#eq-1|(1)]]) by using the same method. Therefore model has a unique solution (Eq.[[#eq-1|(1)]]). |
| − | ==5 Numerical algorithm with ABC fractional derivative== | + | ==5. Numerical algorithm with ABC fractional derivative== |
| − | It is not possible to solve various real and physical applications developed using fractional PDEs accurately. However, a numerical approach to the solution is always enough to take care of a problem in engineering and science. For the Adams-Bashforth-Moulton technique, it can be used here to score a solution. Compared to the RK4, ABMM offers many noteworthy benefits, due to the fact that RK4 calculates four function evaluations per integration step while ABMM only calculates two [ | + | It is not possible to solve various real and physical applications developed using fractional PDEs accurately. However, a numerical approach to the solution is always enough to take care of a problem in engineering and science. For the Adams-Bashforth-Moulton technique, it can be used here to score a solution. Compared to the RK4, ABMM offers many noteworthy benefits, due to the fact that RK4 calculates four function evaluations per integration step while ABMM only calculates two [42]. As a result of the wider interpolation interval, Adams-Moulton methods produce more precise approximations. Generally speaking, implicit procedures are more stable than their explicit counterparts and they also achieve a greater order with the same number of preceding steps. Naturally, its implicit nature makes it challenging to solve because it results in a non-linear equation. |
In this part, we discuss the plant diseases model known as the SPEIR model with the Atangana-Baleanu fractional operator to show the effectiveness, excellence and generality of our approach. All analytical and numerical analyses were detailed throughout the time spent computation using the MATLAB software package. | In this part, we discuss the plant diseases model known as the SPEIR model with the Atangana-Baleanu fractional operator to show the effectiveness, excellence and generality of our approach. All analytical and numerical analyses were detailed throughout the time spent computation using the MATLAB software package. | ||
| Line 1,035: | Line 985: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\varphi \left(t_{n+1}\right)-\varphi \left(t_{n}\right) = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\{\mathcal{F}\left(t_{n},\varphi _{n}\right)-\mathcal{F}\left(t_{n-1},\varphi _{n-1}\right)\right\}+\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\mathcal{F}\left(t,\varphi \left(t\right)\right)\left(t_{n+1}-t\right)^{\upsilon{-1}}dt | + | | style="text-align: center;" | <math>\begin{align} \varphi \left(t_{n+1}\right)-\varphi \left(t_{n}\right) & = \frac{1-\upsilon }{\chi \left(\upsilon \right)}\left\{\mathcal{F}\left(t_{n},\varphi _{n}\right)-\mathcal{F}\left(t_{n-1},\varphi _{n-1}\right)\right\}\\ |
| − | + | &\quad +\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\mathcal{F}\left(t,\varphi \left(t\right)\right)\left(t_{n+1}-t\right)^{\upsilon{-1}}dt\\ | |
| − | + | &\quad -\frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\mathcal{F}\left(t,\varphi \left(t\right)\right)\left(t_{n}-t\right)^{\upsilon{-1}}dt \end{align}</math> | |
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (65) | | style="width: 5px;text-align: right;white-space: nowrap;" | (65) | ||
| Line 1,085: | Line 1,035: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\mathcal{A}_{\upsilon } = \frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t_{n+1}-t\right)^{\upsilon{-1}}\left\{\frac{t-t_{n-1}}{h}\mathcal{F}\left(t_{n},\varphi _{n}\right)-\frac{t-t_{n}}{h}\mathcal{F}\left(t_{n},\varphi _{n}\right)\right\}dt | + | | style="text-align: center;" | <math>\begin{align} \mathcal{A}_{\upsilon } & = \frac{\upsilon }{\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t_{n+1}-t\right)^{\upsilon{-1}}\left\{\frac{t-t_{n-1}}{h}\mathcal{F}\left(t_{n},\varphi _{n}\right)-\frac{t-t_{n}}{h}\mathcal{F}\left(t_{n},\varphi _{n}\right)\right\}dt\\ |
| − | + | & = \frac{\upsilon \mathcal{F}\left(t_{n},\varphi _{n}\right)}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t_{n+1}-t\right)^{\upsilon{-1}}\mathcal{F}\left(t-t_{n-1}\right)dt\\ | |
| − | + | &\quad -\frac{\upsilon \mathcal{F}\left(t_{n-1},\varphi _{n-1}\right)}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\stackrel=0\left(t_{n+1}-t\right)^{\upsilon{-1}}\mathcal{F}\left(t-t_{n-1}\right)dt\\ | |
| − | + | & = \frac{\upsilon \mathcal{F}\left(t_{n},\varphi _{n}\right)}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left\{\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right\}-\frac{\upsilon \mathcal{F}\left(t_{n-1},\varphi _{n-1}\right)}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left\{\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right\}.\end{align} </math> | |
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (69) | | style="width: 5px;text-align: right;white-space: nowrap;" | (69) | ||
| Line 1,113: | Line 1,062: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\varphi \left(t_{n+1}\right) = \varphi \left(t_{n}\right)+\mathcal{F}\left(t_{n},\varphi _{n}\right)\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\} | + | | style="text-align: center;" | <math>\begin{align}\varphi \left(t_{n+1}\right) & = \varphi \left(t_{n}\right)+\mathcal{F}\left(t_{n},\varphi _{n}\right)\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| − | + | &\quad +\mathcal{F}\left(t_{n-1},\varphi _{n-1}\right)\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}. \end{align}</math> | |
| − | + | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (71) | | style="width: 5px;text-align: right;white-space: nowrap;" | (71) | ||
|} | |} | ||
| − | Therefore, the model's solution ([[#eq-1|1]]) is | + | Therefore, the model's solution (Eq.[[#eq-1|(1)]]) is |
{| class="formulaSCP" style="width: 100%; text-align: left;" | {| class="formulaSCP" style="width: 100%; text-align: left;" | ||
| Line 1,127: | Line 1,075: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\begin{ | + | | style="text-align: center;" | <math>\begin{align} \left(S_{1}\right)_{n+1} & = \left(S_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| + | &\quad \left\{r(M-N)-\left(S_{1}\right)_{n}\left(t_{n}\right)\left(\gamma -\frac{a_{1}}{M}\left(I_{1}\right)_{n}\left(t_{n}\right)-\alpha \right)+\beta \left(P_{1}\right)_{n}\left(t_{n}\right)\right\}\\ | ||
| + | &\quad +\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}\\ | ||
| + | &\quad \left\{r(M-N)-\left(S_{1}\right)_{n-1}\left(t_{n-1}\right)\left(\gamma -\frac{a_{1}}{M}\left(I_{1}\right)_{n-1}\left(t_{n-1}\right)-\alpha \right)+\beta \left(P_{1}\right)_{n-1}\left(t_{n-1}\right)\right\}, \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (72) | | style="width: 5px;text-align: right;white-space: nowrap;" | (72) | ||
| Line 1,137: | Line 1,088: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left(P_{1}\right)_{n+1} | + | | style="text-align: center;" | <math>\begin{align}\left(P_{1}\right)_{n+1} & = \left(P_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| − | + | &\quad \left\{\alpha \left(S_{1}\right)_{n}\left(t_{n}\right)+\left(P_{1}\right)_{n}\left(t_{n}\right)\left(\beta{-\gamma}\right)\right\}\\ | |
| − | + | &\quad +\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}\\ | |
| + | &\quad \left\{\alpha \left(S_{1}\right)_{n-1}\left(t_{n-1}\right)+\left(P_{1}\right)_{n-1}\left(t_{n-1}\right)\left(\beta{-\gamma}\right)\right\},\end{align} </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (73) | | style="width: 5px;text-align: right;white-space: nowrap;" | (73) | ||
| Line 1,149: | Line 1,101: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left(E_{1}\right)_{n+1} | + | | style="text-align: center;" | <math>\begin{align}\left(E_{1}\right)_{n+1} & = \left(E_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| − | + | &\quad \left\{\frac{a_{1}}{M}\left(S_{1}\right)_{n}\left(t_{n}\right)\left(I_{1}\right)_{n} \left(t_{n}\right) - \left( \gamma +a_{2} + b_{1}\right)\left(E_{1}\right)_{n}\left(t_{n}\right)\right\}\\ | |
| − | + | &\quad +\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}\\ | |
| + | &\quad \left\{\frac{a_{1}}{M}\left(S_{1}\right)_{n-1}\left(t_{n-1}\right)\left(I_{1}\right)_{n-1}\left(t_{n-1}\right)-\left(\gamma{+}a_{2}+b_{1}\right)\left(E_{1}\right)_{n-1}\left(t_{n-1}\right)\right\}, \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (74) | | style="width: 5px;text-align: right;white-space: nowrap;" | (74) | ||
| Line 1,161: | Line 1,114: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left(I_{1}\right)_{n+1} | + | | style="text-align: center;" | <math>\begin{align}\left(I_{1}\right)_{n+1} & = \left(I_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| − | + | &\quad \left\{a_{2}\left(E_{1}\right)_{n}\left(t_{n}\right)-\left(\gamma{+}a_{3}+b_{2}+\eta \right)\left(I_{1}\right)_{n}\left(t_{n}\right)\right\}\\ | |
| − | + | &\quad +\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}\\ | |
| + | &\quad \left\{a_{2}\left(E_{1}\right)_{n-1}\left(t_{n-1}\right)-\left(\gamma{+}a_{3}+b_{2}+\eta \right)\left(I_{1}\right)_{n-1}\left(t_{n-1}\right)\right\}, \end{align}</math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (75) | | style="width: 5px;text-align: right;white-space: nowrap;" | (75) | ||
| Line 1,173: | Line 1,127: | ||
{| style="text-align: left; margin:auto;width: 100%;" | {| style="text-align: left; margin:auto;width: 100%;" | ||
|- | |- | ||
| − | | style="text-align: center;" | <math>\left(R_{1}\right)_{n+1} = \left(R_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\left\{a_{3}\left(I_{1}\right)_{n}\left(t_{n}\right)-\left(\gamma{+}b_{3}\right)\left(R_{1}\right)_{n}\left(t_{n}\right)\right\} | + | | style="text-align: center;" | <math>\begin{align}\left(R_{1}\right)_{n+1} & = \left(R_{1}\right)_{n}+\left\{\frac{1-\upsilon }{\chi \left(\upsilon \right)}+\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{2ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}\right]-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n}^{\upsilon }}{\upsilon }-\frac{t_{n}^{\upsilon{+1}}}{\upsilon{+1}}\right]\right\}\\ |
| − | + | &\quad \left\{a_{3}\left(I_{1}\right)_{n}\left(t_{n}\right)-\left(\gamma{+}b_{3}\right)\left(R_{1}\right)_{n}\left(t_{n}\right)\right\}\\ | |
| − | + | &\quad +\left\{\frac{\upsilon{-1}}{\chi \left(\upsilon \right)}-\frac{\upsilon }{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\left[\frac{ht_{n+1}^{\upsilon }}{\upsilon }-\frac{t_{n+1}^{\upsilon{+1}}}{\upsilon{+1}}+\frac{t_{n}^{\upsilon{+1}}}{h\chi \left(\upsilon \right)\lambda \left(\upsilon \right)}\right]\right\}\\ | |
| + | &\quad \left\{a_{3}\left(I_{1}\right)_{n-1}\left(t_{n-1}\right)-\left(\gamma{+}b_{3}\right)\left(R_{1}\right)_{n-1}\left(t_{n-1}\right)\right\}.\end{align} </math> | ||
|} | |} | ||
| style="width: 5px;text-align: right;white-space: nowrap;" | (76) | | style="width: 5px;text-align: right;white-space: nowrap;" | (76) | ||
|} | |} | ||
| − | ==6 Numerical simulation | + | ==6. Numerical simulation and discussion== |
| − | + | ||
| − | + | ||
| − | + | This section employs the Adams-Bashforth-Moulton technique to numerically dissolve the fractional operator SPEIR model [40,41]. | |
| + | The values of the initial conditions for model (Eq.[[#eq-1|(1)]]) are given as follows: <math display="inline">M=1000</math>, <math display="inline">S_{1}\left(0\right)=100</math>, <math display="inline">P_{1}\left(0\right)=30</math>, <math display="inline">E_{1}\left(0\right)=60</math>, <math display="inline">I_{1}\left(0\right)=60</math> and <math display="inline">R_{1}\left(0\right)=60</math>. The parameter values applied in the mathematical simulation were extracted from the classical case of the model as shown in [[#table-1|Table 1]] [8,9]. The outcomes of the numerical simulation of the SPEIR model are shown in the paragraphs that follow (with and without control). | ||
| − | + | <div class="center" style="font-size: 75%;">'''Table 1'''. Values of parameters </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | <div id='table-1'></div> | ||
| + | {| class="wikitable" style="margin: 1em auto 0.1em auto;border-collapse: collapse;font-size:85%;width:auto;" | ||
| + | |-style="text-align:center" | ||
| + | | style="text-align:left;" |Parameter | ||
| + | | <math>a_{1}</math> | ||
| + | | <math>a_{2}</math> | ||
| + | | <math>a_{3}</math> | ||
| + | | <math>b_{1}</math> | ||
| + | | <math>b_{2}</math> | ||
| + | | <math>b_{3}</math> | ||
| + | | <math>r</math> | ||
| + | | <math>\alpha </math> | ||
| + | | <math>\beta </math> | ||
| + | | <math>\gamma </math> | ||
| + | | <math>\eta </math> | ||
| + | |- style="text-align:center" | ||
| + | | Value (<math display="inline">\mathrm{\mathcal{R}}_{0}<1</math>) | ||
| + | | 0.06 | ||
| + | | 0.17 | ||
| + | | 0.04 | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | 0.01 | ||
| + | | 0.011 | ||
| + | | 0.0052 | ||
| + | | 0.048 | ||
| + | | 0.004 | ||
| + | | 0.089 | ||
| + | |- style="text-align:center" | ||
| + | | Value (<math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>) | ||
| + | | 0.3 | ||
| + | | 0.17 | ||
| + | | 0.02 | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | 0.01 | ||
| + | | 0.013 | ||
| + | | 0.0052 | ||
| + | | 0.048 | ||
| + | | 0.0008 | ||
| + | | 0.087 | ||
|} | |} | ||
| − | + | ||
| + | [[#img-1|Figure 1]] shows the numerical simulation of plant compartments Susceptible <math display="inline">S_{1}\left(t\right)</math>, Protected <math display="inline">P_{1}\left(t\right)</math> and Latent <math display="inline">E_{1}\left(t\right)</math> with time history for various values of fractional order at <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>. [[#img-1|Figures 1]](a) and [[#img-1|1]](b) show that the maximum value of <math display="inline">S_{1}(t)</math> decreases whether <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>, as the time increases and the fractional-order decreases. [[#img-1|Figures 1]](c) and [[#img-1|1]](d) show that <math display="inline">P_{1}(t)</math> decreases as the fractional order decreases and on a big difference between the situation when <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>. [[#img-1|Figure 1]](e) shows that <math display="inline">E_{1}(t)</math> decreases dramatically during the first period with increasing the fractional order until it reaches about 120 days, and then the process reverses after that. While [[#img-1|Figure 1]](f) shows that <math display="inline">E_{1}(t)</math> is decreasing, but only until 50 days before it starts increasing again. | ||
<div id='img-1'></div> | <div id='img-1'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-1.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 1'''. Numerical simulation of plant compartments <math>S_{1}\left(t\right),P_{1}\left(t\right),E_{1}\left(t\right)</math> for various values of fractional order with <math>\mathrm{\mathcal{R}}_{0}<1</math> and <math>\mathrm{\mathcal{R}}_{0}>1</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| + | |||
| + | |||
| + | [[#img-2|Figure 2]] shows the numerical simulation of plant compartments Infected <math display="inline">I_{1}\left(t\right)</math> and Removed <math display="inline">R_{1}\left(t\right)</math> with time history for several values of fractional order at <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>. [[#img-2|Figure 2]](a) shows that <math display="inline">I_{1}(t)</math> initially decreases until close to 150 days with the value of the fractional order differing, and then the process reverses after that. While [[#img-2|Figure 2]](b) shows that <math display="inline">I_{1}(t)</math> decreases until the period from 60 to 80 days, but its value does not reach zero on the vertical axis, as happened with [[#img-2|Figure 2]](a). | ||
<div id='img-2'></div> | <div id='img-2'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%;" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-2.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 2'''. Numerical simulation of plant compartments <math>I_{1}\left(t\right),R_{1}\left(t\right)</math> for various values of fractional order with <math>\mathrm{\mathcal{R}}_{0}<1</math> and <math>\mathrm{\mathcal{R}}_{0}>1</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| + | |||
| + | |||
| + | [[#img-3|Figure 3]] shows the numerical simulation of plant compartments <math display="inline">S_{1}\left(t\right)</math>, <math display="inline">P_{1}\left(t\right)</math>, <math display="inline">E_{1}\left(t\right)</math>, <math display="inline">I_{1}\left(t\right)</math> and <math display="inline">R_{1}\left(t\right)</math> with time history for various values of fractional order at <math display="inline">\mathrm{\mathcal{R}}_{0}<1</math> and <math display="inline">\mathrm{\mathcal{R}}_{0}>1</math>. | ||
<div id='img-3'></div> | <div id='img-3'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%;" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-3.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 3'''. Numerical simulation of plant compartments <math>S_{1}\left(t\right),P_{1}\left(t\right),E_{1}\left(t\right),I_{1}\left(t\right),R_{1}\left(t\right)</math> with <math>\mathrm{\mathcal{R}}_{0}<1</math> and <math>\mathrm{\mathcal{R}}_{0}>1</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| + | |||
| + | |||
| + | [[#img-4|Figure 4]] shows the dynamic compartments of Susceptible and Infected with various roguing and replanting values at <math display="inline">\upsilon=0.85</math>. [[#img-4|Figures 4]](a) and [[#img-4|4]](b) show that the maximum value of <math display="inline">S_{1}(t)</math> increases as the rate of replanting decreases and the rate of roguing increases. [[#img-4|Figures 4]](c) and [[#img-4|4]](d) show that infectious can be increased as the rate of replanting decreases and the rate of roguing increases. | ||
<div id='img-4'></div> | <div id='img-4'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%;" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-4.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 4'''. Dynamic compartments of Susceptible and Infected with various roguing and replanting values at <math>\upsilon=0.85</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| + | |||
| + | |||
| + | [[#img-5|Figures 5]] and [[#img-6|6]] illustrate the effect of different parameters on plant compartments of americanthe SPEIR model at <math display="inline">\upsilon=0.85</math>. | ||
<div id='img-5'></div> | <div id='img-5'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%;" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-5.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 5'''. The effect of alteration the values of parameters <math>a_{1}</math>, <math>a_{2}</math>, <math>b_{1}</math> and <math>b_{2}</math> on all compartment stages where the fractional operator <math>\upsilon=0.85</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| + | |||
<div id='img-6'></div> | <div id='img-6'></div> | ||
| − | {| class=" | + | {| class="wikitable" style="margin: 0em auto 0.1em auto;border-collapse: collapse;width:75%;" |
| + | |-style="background:white;" | ||
| + | |style="text-align: center;padding:10px;"| [[Image:Draft_Hagag_877846790-6.png|600px]] | ||
|- | |- | ||
| − | | | + | | style="background:#efefef;text-align:left;padding:10px;font-size: 85%;"| '''Figure 6'''. The effect of alteration the values of parameters <math>\alpha </math>, <math>\beta </math>, <math>\gamma </math> and <math>\eta </math> on all compartment stages where the fractional operator <math>\upsilon=0.85</math> |
| − | + | ||
| − | + | ||
|} | |} | ||
| − | ==7 Conclusion== | + | ==7. Conclusion== |
In this study, we considered and analyzed the SPEIR model displayed the dynamics of plant diseases with the Atangana-Baleanu fractional derivative in Caputo sense. Both the NEE and EE points were analyzed in terms of model equilibria and stability analysis (local-global). We also demonstrated the generic form of <math display="inline">\mathrm{\mathcal{R}}_{0}</math> and the effects of the controls proposed on it. The Adams-Bashforth-Moulton approach was used to study and solve numerical simulations of the suggested model. The value of the fractional order <math display="inline">\upsilon </math> as well as the parameters of the SPEIR model affect the numerical results that are obtained. Because of this, solutions generated by the fractional order model typically converge extremely quickly to real issues. | In this study, we considered and analyzed the SPEIR model displayed the dynamics of plant diseases with the Atangana-Baleanu fractional derivative in Caputo sense. Both the NEE and EE points were analyzed in terms of model equilibria and stability analysis (local-global). We also demonstrated the generic form of <math display="inline">\mathrm{\mathcal{R}}_{0}</math> and the effects of the controls proposed on it. The Adams-Bashforth-Moulton approach was used to study and solve numerical simulations of the suggested model. The value of the fractional order <math display="inline">\upsilon </math> as well as the parameters of the SPEIR model affect the numerical results that are obtained. Because of this, solutions generated by the fractional order model typically converge extremely quickly to real issues. | ||
| − | + | '''Conflict of Interest:''' The author declare that there is no Conflict of Interest. | |
| − | === | + | ==Acknowledgments== |
| + | |||
| + | This research project was funded by the Deanship of Scientific Research, Princess Nourah bint Abdulrahman University, through the Program of Research Project Funding After Publication, grant No (43- PRFA-P-19). | ||
| + | |||
| + | ==References== | ||
| + | <div class="auto" style="text-align: left;width: auto; margin-left: auto; margin-right: auto;font-size: 85%;"> | ||
<div id="cite-1"></div> | <div id="cite-1"></div> | ||
| − | + | [1] Vanderplank J.E. Plant disease: epidemics and control. New York, Academic Press, 1963. | |
<div id="cite-2"></div> | <div id="cite-2"></div> | ||
| − | + | [2] Verhoeff K. Latent infection by fungi. Annu Rev Phytopathol, 12:99-107, 1974. | |
<div id="cite-3"></div> | <div id="cite-3"></div> | ||
| − | + | [3] Agrios G.N. Lant pathology. Academic Press, San Diego, 1988. | |
<div id="cite-4"></div> | <div id="cite-4"></div> | ||
| − | + | [4] Zhang T., Meng X., Song Y., Li Z. Dynamical analysis of delayed plant disease models with continuous or impulsive cultural control strategies. Abstract and Applied Analysis, 2012(SI12):1-25, 2012. | |
<div id="cite-5"></div> | <div id="cite-5"></div> | ||
| − | + | [5] Sisterson M.S., Stenger D.C. Roguing with replacement in perennial crops: conditions for successful disease management. Phytopathology, 103:117-28, 2013. | |
<div id="cite-6"></div> | <div id="cite-6"></div> | ||
| − | + | [6] Yang W., Wang D., Yang L. A stable numerical method for space fractional LandauLifshitz equations. Applied Mathematics Letters, 61:149-155, 2016. | |
<div id="cite-7"></div> | <div id="cite-7"></div> | ||
| − | + | [7] Kuang Y., Hu G. An adaptive FEM with ITP approach for steady Schrödinger equation. International Journal of Computer Mathematics, 95:187-201, 2018. | |
<div id="cite-8"></div> | <div id="cite-8"></div> | ||
| − | + | [8] Luo Y., Gao S., Xie D., Dai Y. A discrete plant disease model with roguing and replanting. Advances in Difference Equations, 1:1-18, 2015. | |
<div id="cite-9"></div> | <div id="cite-9"></div> | ||
| − | + | [9] Chan M.S., Jeger M.J. An analytical model of plant virus disease dynamics with roguing and replanting. Journal of Applied Ecology, 31:413-427, 1994. | |
<div id="cite-10"></div> | <div id="cite-10"></div> | ||
| − | + | [10] Blas N., David G. Dynamical roguing model for controlling the spread of tungro virus via Nephotettix Virescens in a rice field. Journal of Physics: Conference Series, 893, 012018, 2017. | |
<div id="cite-11"></div> | <div id="cite-11"></div> | ||
| − | + | [11] Anggriani N., Yusuf M., Supriatna A.K. The effect of insecticide on the vector of rice tungro disease: insight from a mathematical model. International Information Institute (Tokyo), 20:6197-206, 2017. | |
<div id="cite-12"></div> | <div id="cite-12"></div> | ||
| − | + | [12] Anggriani N., Istifadah N., Hanifah M., Supriatna A.K. A mathematical model of protectant and curative fungicide application and its stability analysis. IOP Conference Series: Earth and Environmental Science, 31, 012014, 2016. | |
<div id="cite-13"></div> | <div id="cite-13"></div> | ||
| − | + | [13] Anggriani N., et al. Optimal control of plant disease model with roguing, replanting, curative, and preventive treatment. In Journal of Physics: Conference Series, 1657, 012050, 2020. | |
<div id="cite-14"></div> | <div id="cite-14"></div> | ||
| − | + | [14] Anggriani N., et al. Dynamical analysis of plant disease model with roguing, replanting and preventive treatment. In Proceedings of 4th International Conferenve on Research, Implementation, and Education of Mathematics and Science, Yogyakarta, 14–16 May 2017. | |
<div id="cite-15"></div> | <div id="cite-15"></div> | ||
| − | + | [15] Odibat Z., Kumar S. A robust computational algorithm of homotopy asymptotic method for solving systems of fractional differential equations. Journal of Computational and Nonlinear Dynamics, 14, 081004, 2019. | |
| + | |||
| + | <div id="cite-15"></div> | ||
| + | [16] Momani S., Abu Arqub O., Maayah B. Piecewise optimal fractional reproducing kernel solution and convergence analysis for the Atangana–Baleanu–Caputo model of the Lienard’s equation. Fractals, 28, 2040007, 2020. | ||
<div id="cite-16"></div> | <div id="cite-16"></div> | ||
| − | + | [17] Akgul A., Cordero A., Torregrosa J.R. Solutions of fractional gas dynamics equation by a new technique. Mathematical Methods in the Applied Sciences, 43:1349-1358, 2020. | |
<div id="cite-17"></div> | <div id="cite-17"></div> | ||
| − | + | [18] Singh J., Swroop R., Kumar D. A computational approach for fractional convection-diffusion equation via integral transforms. Ain Shams Engineering Journal, 9:1019-1028, 2018. | |
<div id="cite-18"></div> | <div id="cite-18"></div> | ||
| − | + | [19] Caputo M., Fabrizio M. A new definition of fractional derivative without singular kernel. Progress in Fractional Differentiation and Applications, 1:73-85, 2015. | |
<div id="cite-19"></div> | <div id="cite-19"></div> | ||
| − | + | [20] Abu Arqub O., Singh J., Maayah B., Alhodaly M. Reproducing kernel approach for numerical solutions of fuzzy fractional initial value problems under the Mittag–Leffler kernel differential operator. Mathematical Methods in the Applied Sciences, 46(7):7965-7986, 2023. | |
| + | |||
| + | <div id="cite-19"></div> | ||
| + | [21] Baleanu D., Mousalou A., Rezapour S. On the existence of solutions for some infinite coefficient-symmetric Caputo-Fabrizio fractional integro-differential equations. Boundary Value Problems, 1:1-9, 2017. | ||
<div id="cite-20"></div> | <div id="cite-20"></div> | ||
| − | + | [22] Teodoro G., et al. A review of definitions of fractional derivatives and other operators. Journal of Computational Physics, 388:195-208, 2019. | |
| + | |||
| + | <div id="cite-21"></div> | ||
| + | [23] Atangana A., Baleanu D. New fractional derivatives with nonlocal and non-singular kernel: theory and application to heat transfer model. Thermal Science, 20:763-769, 2016. | ||
| + | |||
| + | <div id="cite-21"></div> | ||
| + | [24] Maayah B., Abu Arqub O., Alnabulsi S., Alsulami H. Numerical solutions and geometric attractors of a fractional model of the cancer-immune based on the Atangana-Baleanu-Caputo derivative and the reproducing kernel scheme. Chinese Journal of Physics, 80:463-483, 2022. | ||
| + | |||
| + | <div id="cite-21"></div> | ||
| + | [25] Abu Arqub O., Singh J., Alhodaly M. Adaptation of kernel functions‐based approach with Atangana–Baleanu–Caputo distributed order derivative for solutions of fuzzy fractional Volterra and Fredholm integrodifferential equations. Mathematical Methods in the Applied Sciences, 46:7807-7834, 2023. | ||
<div id="cite-21"></div> | <div id="cite-21"></div> | ||
| − | + | [26] Djennadi S., Shawagfeh N., Abu Arqub O. Well-posedness of the inverse problem of time fractional heat equation in the sense of the Atangana-Baleanu fractional approach. Alexandria Engineering Journal, 59:2261-2268, 2020. | |
<div id="cite-22"></div> | <div id="cite-22"></div> | ||
| − | + | [27] Akgül A. A novel method for a fractional derivative with nonlocal and non-singular kernel. Chaos, Solitons Fractals, 114:478-482, 2018. | |
<div id="cite-23"></div> | <div id="cite-23"></div> | ||
| − | + | [28] Jajarmi A., Ghanbari B., Baleanu D. A new and efficient numerical method for the fractional modelling and optimal control of diabetes and tuberculosis co-existence. Chaos: An Interdisciplinary Journal of Nonlinear Science 29, 093111, 2019. | |
<div id="cite-24"></div> | <div id="cite-24"></div> | ||
| − | + | [29] Yildiz T.A., et al. New aspects of time fractional optimal control problems within operators with nonsingular kernel. Discrete & Continuous Dynamical Systems-S, 13:407-28, 2020. | |
<div id="cite-25"></div> | <div id="cite-25"></div> | ||
| − | + | [30] Sardar T., et al. A generic model for a single strain mosquito-transmitted disease with memory on the host and the vector. Mathematical Biosciences, 263:18-36, 2015. | |
<div id="cite-26"></div> | <div id="cite-26"></div> | ||
| − | + | [31] Podlubny I. Fractional differential equations. Academic Press, New York (1999). | |
<div id="cite-27"></div> | <div id="cite-27"></div> | ||
| − | + | [32] Gorenflo R., Mainardi F. Fractional calculus: Int and differential equations of fractional order. In A. Carpinteri, F. Mainardi (Eds.), Fractals and Fractional Calculus, New York, 1997. | |
<div id="cite-28"></div> | <div id="cite-28"></div> | ||
| − | + | [33] Driessche P., Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathematical Biosciences, 180:29-48, 2002. | |
<div id="cite-29"></div> | <div id="cite-29"></div> | ||
| − | + | [34] Castillo-Chavez C., Feng Z., Huang W. On the computation of R0 and its role in global stability. IMA Volumes in Mathematics and Its Applications, 125:229-250, 2002. | |
<div id="cite-30"></div> | <div id="cite-30"></div> | ||
| − | + | [35] Diekmann O., Heesterbeek J.A.P. Mathematical epidemiology of infectious diseases: Model building, analysis and interpretation. John Wiley & Sons, Vol. 5, pp. 320, 2000. | |
<div id="cite-31"></div> | <div id="cite-31"></div> | ||
| − | + | [36] Nise N.S. Control systems engineering. John Wiley & Sons, 8th edition, pp. 800, 2020. | |
<div id="cite-32"></div> | <div id="cite-32"></div> | ||
| − | + | [37] Verhulst F. Nonlinear differential equations and dynamical systems. Springer, Heidelberg, pp. 306, 1996. | |
<div id="cite-33"></div> | <div id="cite-33"></div> | ||
| − | + | [38] Wiggins S. Introduction to applied nonlinear dynamical systems and chaos. Part of the book series: Texts in Applied Mathematics (TAM, volume 2), 2nd Edition, Springer-Verlag, Heidelberg, pp. 882, 2003. | |
<div id="cite-34"></div> | <div id="cite-34"></div> | ||
| − | + | [39] Kumar D., Singh J., Baleanu D. Analysis of regularized long-wave equation associated with a new fractional operator with Mittag-Leffler type kernel. Physica A, 492:155-167, 2018. | |
<div id="cite-35"></div> | <div id="cite-35"></div> | ||
| − | + | [40] Diethelm K., Freed A.D. The FracPECE subroutine for the numerical solution of differential equations of fractional order. Forschung und wissenschaftliches Rechnen, 57-71, 1999. | |
<div id="cite-36"></div> | <div id="cite-36"></div> | ||
| − | + | [41] Garrappa Z.R. On linear stability of predictor-corrector algorithms for fractional differential equations. International Journal of Computer Mathematics, 87:2281-2290, 2010. | |
<div id="cite-37"></div> | <div id="cite-37"></div> | ||
| − | + | [42] Butcher J. Numerical methods for ordinary differential equations in the 20th century. Journal of Computational and Applied Mathematics, 125:1-29, 2000. | |
Latest revision as of 09:41, 21 July 2023
Abstract
Food security has become a significant issue due to the growing human population. In this case, a significant role is played by agriculture. The essential foods are obtained mainly from plants. Plant diseases can, however, decrease both food production and its quality. Therefore, it is substantial to comprehend the dynamics of plant diseases as they can provide insightful information about the dispersal of plant diseases. In order to investigate the dynamics of plant disease and analyze the effects of strategies of disease control, a mathematical model can be applied. We show that this model provides the non-negative solutions that population dynamics requires. The model was investigated by using the Atangana-Baleanu in Caputo sense (ABC) operator which is symmetrical to the Caputo-Fabrizio (CF) operator with a different function. Whereas the ABC operator uses the generalized Mittag-Leffler function while the CF operator employs the exponential kernel. For the proposed model, we have displayed the local and global stability of a nonendemic and an endemic equilibrium, existence and uniqueness theorems. By applying the fractional Adams-Bashforth-Moulton method, we have implemented numerical solutions to illustrate the theoretical analysis.
Keywords: Adams-Bashforth-Moulton method, local and global asymptotic equilibrium stability, sensitivity analysis, existence theorems, uniqueness theorems, plant diseases model, numerical simulations
1. Introduction
Plants are an incredibly valuable component of our world. The earth, due to the presence of plants, is known as a green planet. They are perhaps the most important component of the life of all the earth's living beings. Some of the plant's essential functions are food, reducing the level of pollution, supplying fresh oxygen, medications, furniture and refuge. Plant disease, however, triggers a decline in food production and efficiency, which can lead to numerous health and social issues. Moreover, it can cause considerable economic ramifications.
Plant disease epidemiology studies the evolution of populations of plant diseases in time and space. Usually, the execution of the techniques is accomplished by roguing and then replanting, which offers two possible advantages. Initially, infected plants could be removed and could inoculum sources could be reduced, possibly slowing the dispersal of pathogens. Then, the infected plant may die or suffer a reduction in yield. Consequently, substituting diseased plants with healthful plants can indemnity crop casualties. The exposed plant can also be able to spread disease in some cases [1-3]. Since the exposed plant is capable of spreading disease, the propagation of plant disease can be more rapid. It is therefore important to realize the impact of plants uncovered on the dynamics of plant disease contamination. Their simulation revealed that the application of fungicides is efficient in minimizing population infection [4-6].
Mathematical modeling is useful in explaining how diseases dispersal and different factors involved in the dispersal of the disease have been specified [7-11]. The defensive and curative fungicide model was introduced in Anggriani et al. [12], where it has been split into three ingredients: Infectious, protected and susceptible. Their simulation revealed that the implementation of fungicides is active in minimizing population infection. Model plant diseases with replanting, roguing and preventive care have been introduced in Anggriani et al. [13] without consideration to curative therapy. In 2017, Anggriani et al. [14] created a plant disease mathematical model that includes five ingredients: Susceptible, Protected, Infectious, Exposed and Post-Infectious with protective curative therapies. They observed that by using curative and preventative care, the transmission of plant disease can be minimized. However, where only one therapy is offered, preventive therapy is favored over curative therapy.
In actuality, there are various meanings of fractional derivatives which in general do not necessarily correspond. One of these definitions which is often utilized in different implementations of fractional differential equations is Caputo fractional derivative [15-18]. There is also a novel fractional derivative definition, named the Caputo-Fabrizio fractional operator, which centered on the exponential function [19-21]. And in the same context, there is a novel fractional derivative definition, named the Atangana-Baleanu fractional operator, which centered on the Mittag-Leffler function [22-27]. Many authors have successfully attempted to model actual processes utilizing this fractional derivative operator [28-29].
We consider a plant disease spread model within fractional calculus, where a susceptible person crosses an exposed stage prior to becoming an infectious person and diseases may also be passed on by exposed plants. The major purpose for this protraction is that the plant diseases of the classical case [12-14] do not load any acquaintance about the memory and learning techniques that impact the propagation of a disease [25]. Now, we regard the plant diseases model with fractional order as follows:
|
|
(1) |
With initial conditions , , , and , where indicate to the overall population of the actual plant, is representing the Susceptible population, is representing the Protected population, is representing the Exposed population, is representing the Infectious/Removed population, is representing the Recovered population, and , this suggests that the size of population is not steady. (i.e. size of the population is variable). The actual meaning of each of the model's parameters, all of which have positive values, is as follows:
- : rate of replanting.
- : disease progression diversion rate for latent compartment.
- : disease progression diversion rate for infected compartment.
- : disease progression diversion rate for removed compartment.
- : overall maximum plant population (agronomy).
- : influence accumulative death rate for latent compartment.
- : influence accumulative death rate for infected compartment.
- : influence accumulative death rate for latent removed compartment.
- : efficacy preventive therapy.
- : preventive therapy rate.
- : rate of natural death.
- : rate of roguing.
Some assumptions have been made to facilitate understanding this model [american14]:
- There is no closure of the plant population due to replanting and natural death.
- The infected plant compartment comprises of two compartments, called, latent and infected .
- The infected plant is lifted if it shows comprehensive symptoms.
- The insect vector and environment factor are neglected.
- The Preventive therapy (insecticide) be presented to susceptible compartments.
- The Protected compartment contains the Susceptible plants that have received protective therapy.
- Protected plants have defensive or prevention impact, but are not immune to disease, therefore it is permitting re-entry into the susceptible compartment.
This article is structured to have some significant preliminaries in Section 2. Section 3 transacts with studying the local and global asymptotic equilibrium stability (the disease-free case and the endemic case) and the sensibility analysis of reproduction number () without control of our model (Eq.(1)). Section 4 transacts with studying the existence and uniqueness theorems of our model (Eq.(1)). Then, computational technique (Adams-Bashforth-Moulton method) are graphically represented and covered in Sections 5 and 6. Finally, conclusions are drawn.
2. Preliminaries
Recently, many fractional calculus concepts and definitions have been developed [31-32].
Definition 1
The fractional derivative of the Atangana-Baleanu in Caputo sense (ABC) is denoted as
|
|
(2) |
where is a normalized function with which are symmetrical to the Caputo-Fabrizio (CF) case, , , , where is the Sobolev space of order 1 in and indicate to a Mittag-Leffler function expressed as
|
|
(3) |
Definition 2
The related fractional integral to the AB Caputo operator is denoted as
|
|
(4) |
3. Analysis of plant disease model
3.1 The local asymptotic equilibrium stability
Since , , , and when , we can use them to find two equilibrium points of the fractional system (Eq.(1)), by solving the above equations we get
3.1.1 Non endemic equilibrium (NEE) point
The NEE solution of the system (Eq.(1)) is
|
|
(5) |
with where , , . The basic reproduction number that is calculated utilizing the generation operator method [28,29] can be found in [14] as follows:
|
|
(6) |
It is known as the number of secondary contagions induced by a single primary contagion in a completely susceptible population [35] and is generally expressed using model (Eq.(1)). The rate of the basic reproduction number counts on the replanting rate value. This is the justification that we should replant the plant if we want to control plant disease.
3.1.2 Endemic equilibrium (EE) point
The endemic equilibrium solution of the system (Eq.(1)) is
|
|
where
|
|
(7) |
with . Conclusion of the outcomes of the system's equilibrium points (Eq.(1)) with prepared to research analytically the stability of the equilibrium points.
Theorem 3.1.2.1. The NEE point of model (Eq.(1)) is locally asymptotically stable if and unstable if .
Proof. The system's Jacobian matrix (Eq.(1)) at is
|
|
(8) |
The eigenvalues of are given as and the following quadratic equation gives and as
|
|
(9) |
From Eq.(9), we can notice that if , then is locally asymptotically stable. This suggests that all polynomial coefficients (9) have the same signal, then the eigenvalues (roots) have negative real part. But if , the NEE is unstable and this would lead to a stable endemic equilibrium being present of . Now by proving the theorem of local stability of , we conclude this section.
Theorem 3.1.2.2. If , the endemic equilibrium of model (Eq.(1)) is locally asymptotically stable.
Proof. The system's Jacobian matrix (Eq.(1)) at is
|
|
(10) |
and its polynomial characteristic written as
|
|
(11) |
The eigenvalues of are and the roots of the polynomial . By applying the Routh-Hortwitz criterian [31] we find that , and . Therefore every the polynomial roots have a negative real part when [32,33]. This implies that is locally asymptotically stable when [30].
3.2 The global asymptotic equilibrium stability
Theorem 3.2.1. The disease-free equilibrium of the plant disease model is globally asymptotically stable in the suitable range if and unstable if .
Proof. We are applying the Lyapunov function [33], which is defined by
|
|
(12) |
where
|
|
(13) |
englishConsequently, its derivative along the plant disease model's solutions
|
|
(14) |
|
|
(15) |
|
|
(16) |
Now we divide by S to get
|
|
(17) |
Since is major than and categories, so
|
|
(18) |
Since . According to the proposed model, plant disease model would therefore be eliminated if and only if . In general, because all parameters are positive in the plant disease model, Lyapunov function therefore decreases if and increases if , eventually if . is therefore the function of Lyapunov within the practicable biological interval and the greater compact invariant set in is the point . Every solution of the plant disease model proposed in this study with an initial term in tends to when if and only if through the well-known Lasalles invariance principle [38]. In conclusion, the plant disease model's disease-free equilibrium presented here is globally asymptotically stable.
Theorem 3.2.2. The endemic equilibrium point of the plant disease system is globally asymptotically stable if .
Proof. We use the Lyapunov function [38] to prove this
|
|
(19) |
Consequently, applying the derivative to both sides gives
|
|
(20) |
replacing , , , and by their values, we obtain
|
|
(21) |
Then we have
|
|
(22) |
They can be separated in two part as follows
|
|
(23) |
|
|
(24) |
This can be simplified as
|
|
(25) |
where
|
|
(26) |
and
|
|
(27) |
this implies if , if and if , this implies , , , and .
We can now conclude that the largest compact invariant set for the plant diseases model in is the point the endemic equilibrium of the plant diseases model.
3.3 Sensibility analysis of reproduction number ( ) without control3.3 Sensibility analysis of reproduction number (
R
0
{\displaystyle \mathrm {\mathcal {R}} {0}}
) without control
Since the parameters of the epizootic system are either predestined or equipped, this raises some doubt as to their values used to draw a conclusion about the underlying epidemic. Therefore, it's critical to identify the specific effects of each element on the dynamics of the pestilence and group the variables that have the most impact to limit or spread the outbreak. Within this section, a sensitivity analysis is conducted for the disease parameters identified with the suggested SPEIR model via the sensitivity indicator. Quantifying the most sensitive aspects of the basal reproductive number can be done with the help of the Sensitivity Indicator strategy. The following equations give the standardized sensitivity indicator of for all the parameters used within the SPEIR model in Table 1 where
|
|
(28) |
As shown in the previous calculations, some component of the sensitivity indicator are positive, like , , and , while others, like , , and are negative. Furthermore, the most important feature of these indicators is the functionality of the SPEIR model parameters. This means that getting a small amendment in one of the parameters will amendment the epidemic dynamics. The value displays that decreasing (increasing) for example by 70% increases (decreases) the basic reproductive number by about 70% A small change in a parameter can head to comparatively enormous quantitative changes, requiring these sensitive parameters to be understood. It can be shown from previous calculations that parameters (disease progression diversion rate for Latent Compartmentenglish) and (rate of roguingenglish) are, respectively, the maximum and minimum sensitivity epidemical parameters .
4. Achieve existence and uniqueness
In this section, we will prove that model 4 has a unique solution, that the kernel satisfies Lipschitz's condition and that the functions in this model is bounded.
Now, we will analyze the fractional model (Eq.(1)). Usage of an integral fractional operator on Eq. (1), we are gaining
|
where
|
|
(30) |
When has an upper limit, the Lipschitz condition for the will be fulfilled. So, if has an upper limit, we find that
|
|
(31) |
that is and , where and are bounded functions and . We have that,
|
|
similarly, we obtain the other kernels as following
|
where
|
|
(36) |
and , , , , , , , . Hence, for the kernels , , , and , the Lipschitz condition is justified.
Theorem 4.1. Presume that is obliged, then the operator is supplied by
|
|
(37) |
satisfies the Lipschitz condition.
Proof. Assume that and are bounded functions with , then we have
|
|
(38) |
This completes the proof. The same process can be applied to , , and .
Theorem 4.2. If is a bounded, then the operator
|
|
(39) |
satisfies
(i)
|
|
(40) |
where is the inner product space bounded in .
(ii)
|
|
(41) |
Proof(i). Suppose that is bounded function, then
|
|
(42) |
Proof(ii). Suppose that , since is bounded function, so
|
|
(43) |
This completes the proof. The same process can be applied to , , and .
An inquiry into the existence and uniqueness of Eq.(1) will be discussed in the following. From Eq. (29) we can write
|
|
(44) |
The difference of the successive term can be written as
|
|
(45) |
According to Castillo-Chavez et al. [34], it would be easy to write
|
|
(46) |
Then, from Eq. (45), we get
|
|
(47) |
By triangular inequality, the above Eq. turn into
|
|
(48) |
Whereas, the kernel fulfilled the Lipschitz condition. we have
|
|
(49) |
Theorem 4.3. If fits the following condition
|
|
(50) |
then, model (Eq.(1)) has a unique solution.
Proof. Suppose that is bounded. As the Lipschitz condition is fulfilled by the kernel, therefore, utilizing the recursive process of Eq. (49), we acquire
|
|
Therefore, the function offered by Eq. (46) exists and is smooth as well. Currently, we wish to illustrate that the above-mentioned functions are actually a solution to englishmodel (Eq.(1)). Presume that
|
|
(51) |
So that,
|
|
(52) |
Using the recursive approach once more, we get
|
|
(53) |
If , we get
|
|
(54) |
Using the limit in Eq. (54) as gets closer to , we arrive at . Thus, the existence is demonstrated. It is still necessary to demonstrate the uniqueness of the model english (Eq.(1)). Assume that is another solution of model (Eq.(1)), then
|
|
(55) |
Using the norm for Eq. (55), we get
|
|
(56) |
Because the kernel fulfills the Lipschitz condition, thus we may write
|
|
(57) |
Consequently,
|
|
(58) |
If
|
|
(59) |
then,
|
|
(60) |
Thus, the uniqueness is verified. We can prove the uniqueness of the rest of equations in model (Eq.(1)) by using the same method. Therefore model has a unique solution (Eq.(1)).
5. Numerical algorithm with ABC fractional derivative
It is not possible to solve various real and physical applications developed using fractional PDEs accurately. However, a numerical approach to the solution is always enough to take care of a problem in engineering and science. For the Adams-Bashforth-Moulton technique, it can be used here to score a solution. Compared to the RK4, ABMM offers many noteworthy benefits, due to the fact that RK4 calculates four function evaluations per integration step while ABMM only calculates two [42]. As a result of the wider interpolation interval, Adams-Moulton methods produce more precise approximations. Generally speaking, implicit procedures are more stable than their explicit counterparts and they also achieve a greater order with the same number of preceding steps. Naturally, its implicit nature makes it challenging to solve because it results in a non-linear equation.
In this part, we discuss the plant diseases model known as the SPEIR model with the Atangana-Baleanu fractional operator to show the effectiveness, excellence and generality of our approach. All analytical and numerical analyses were detailed throughout the time spent computation using the MATLAB software package.
Taking into account the following fractional differential equation
|
|
(61) |
We apply the basic calculus theorem in order to transform the Eq. (61) to
|
|
(62) |
at we find that
|
|
(63) |
and at , we have
|
|
(64) |
The result of subtracting (64) from (63) is as follows
|
|
(65) |
Consequently,
|
|
(66) |
where
|
|
(67) |
and
|
|
(68) |
Then, we have
|
|
(69) |
likewise, we obtain
|
|
(70) |
The analytical solution is thus given as
|
|
(71) |
Therefore, the model's solution (Eq.(1)) is
|
|
(72) |
|
|
(73) |
|
|
(74) |
|
|
(75) |
|
|
(76) |
6. Numerical simulation and discussion
This section employs the Adams-Bashforth-Moulton technique to numerically dissolve the fractional operator SPEIR model [40,41].
The values of the initial conditions for model (Eq.(1)) are given as follows: , , , , and . The parameter values applied in the mathematical simulation were extracted from the classical case of the model as shown in Table 1 [8,9]. The outcomes of the numerical simulation of the SPEIR model are shown in the paragraphs that follow (with and without control).
| Parameter | |||||||||||
| Value () | 0.06 | 0.17 | 0.04 | 0 | 0 | 0.01 | 0.011 | 0.0052 | 0.048 | 0.004 | 0.089 |
| Value () | 0.3 | 0.17 | 0.02 | 0 | 0 | 0.01 | 0.013 | 0.0052 | 0.048 | 0.0008 | 0.087 |
Figure 1 shows the numerical simulation of plant compartments Susceptible , Protected and Latent with time history for various values of fractional order at and . Figures 1(a) and 1(b) show that the maximum value of decreases whether and , as the time increases and the fractional-order decreases. Figures 1(c) and 1(d) show that decreases as the fractional order decreases and on a big difference between the situation when and . Figure 1(e) shows that decreases dramatically during the first period with increasing the fractional order until it reaches about 120 days, and then the process reverses after that. While Figure 1(f) shows that is decreasing, but only until 50 days before it starts increasing again.
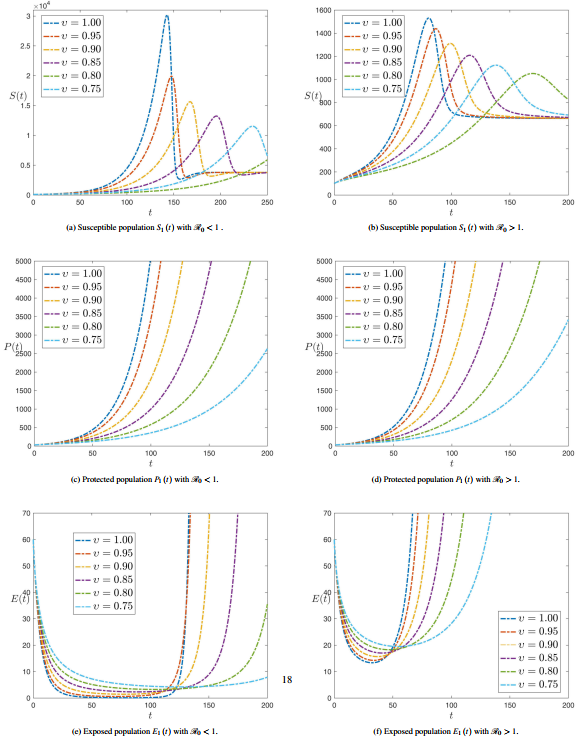
|
| Figure 1. Numerical simulation of plant compartments for various values of fractional order with and |
Figure 2 shows the numerical simulation of plant compartments Infected and Removed with time history for several values of fractional order at and . Figure 2(a) shows that initially decreases until close to 150 days with the value of the fractional order differing, and then the process reverses after that. While Figure 2(b) shows that decreases until the period from 60 to 80 days, but its value does not reach zero on the vertical axis, as happened with Figure 2(a).
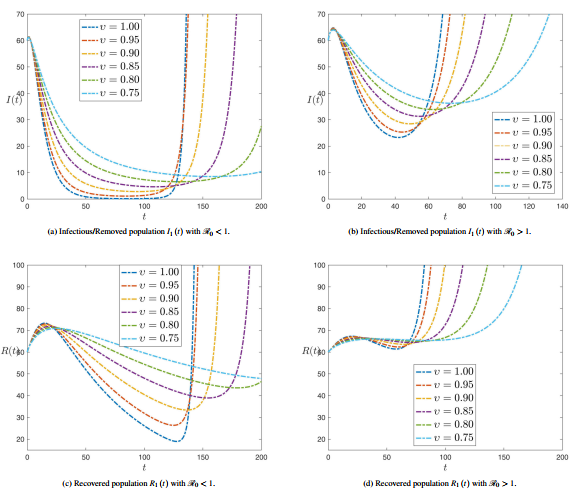
|
| Figure 2. Numerical simulation of plant compartments for various values of fractional order with and |
Figure 3 shows the numerical simulation of plant compartments , , , and with time history for various values of fractional order at and .
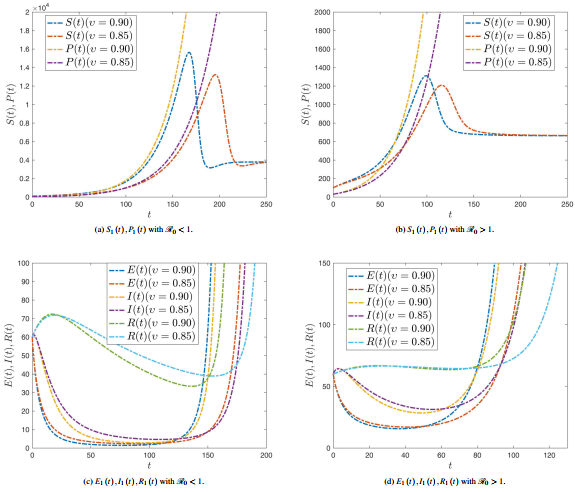
|
| Figure 3. Numerical simulation of plant compartments with and |
Figure 4 shows the dynamic compartments of Susceptible and Infected with various roguing and replanting values at . Figures 4(a) and 4(b) show that the maximum value of increases as the rate of replanting decreases and the rate of roguing increases. Figures 4(c) and 4(d) show that infectious can be increased as the rate of replanting decreases and the rate of roguing increases.
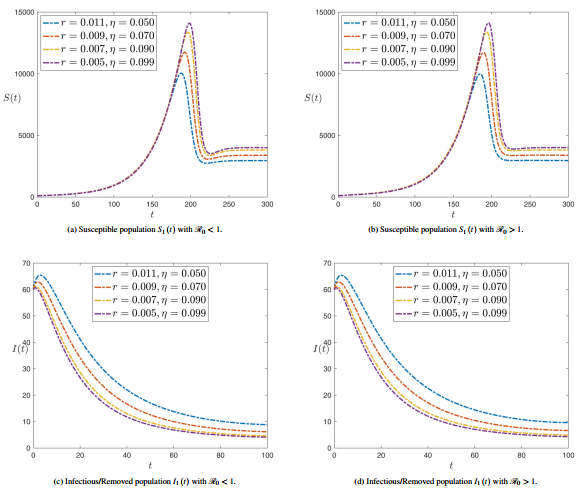
|
| Figure 4. Dynamic compartments of Susceptible and Infected with various roguing and replanting values at |
Figures 5 and 6 illustrate the effect of different parameters on plant compartments of americanthe SPEIR model at .
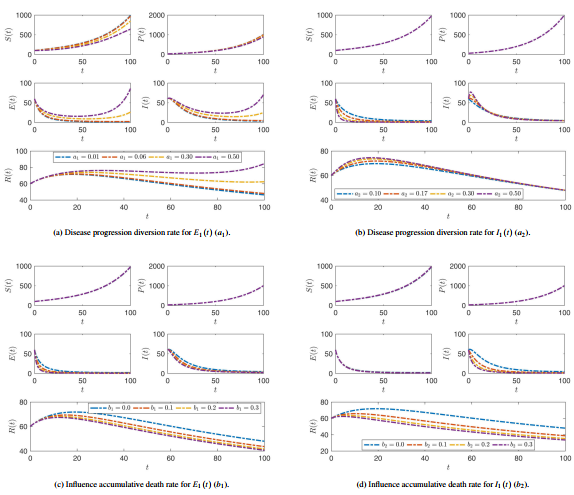
|
| Figure 5. The effect of alteration the values of parameters , , and on all compartment stages where the fractional operator |
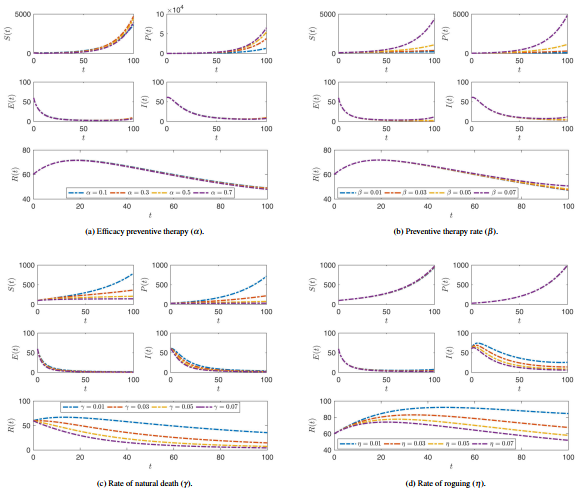
|
| Figure 6. The effect of alteration the values of parameters , , and on all compartment stages where the fractional operator |
7. Conclusion
In this study, we considered and analyzed the SPEIR model displayed the dynamics of plant diseases with the Atangana-Baleanu fractional derivative in Caputo sense. Both the NEE and EE points were analyzed in terms of model equilibria and stability analysis (local-global). We also demonstrated the generic form of and the effects of the controls proposed on it. The Adams-Bashforth-Moulton approach was used to study and solve numerical simulations of the suggested model. The value of the fractional order as well as the parameters of the SPEIR model affect the numerical results that are obtained. Because of this, solutions generated by the fractional order model typically converge extremely quickly to real issues.
Conflict of Interest: The author declare that there is no Conflict of Interest.
Acknowledgments
This research project was funded by the Deanship of Scientific Research, Princess Nourah bint Abdulrahman University, through the Program of Research Project Funding After Publication, grant No (43- PRFA-P-19).
References
[1] Vanderplank J.E. Plant disease: epidemics and control. New York, Academic Press, 1963.
[2] Verhoeff K. Latent infection by fungi. Annu Rev Phytopathol, 12:99-107, 1974.
[3] Agrios G.N. Lant pathology. Academic Press, San Diego, 1988.
[4] Zhang T., Meng X., Song Y., Li Z. Dynamical analysis of delayed plant disease models with continuous or impulsive cultural control strategies. Abstract and Applied Analysis, 2012(SI12):1-25, 2012.
[5] Sisterson M.S., Stenger D.C. Roguing with replacement in perennial crops: conditions for successful disease management. Phytopathology, 103:117-28, 2013.
[6] Yang W., Wang D., Yang L. A stable numerical method for space fractional LandauLifshitz equations. Applied Mathematics Letters, 61:149-155, 2016.
[7] Kuang Y., Hu G. An adaptive FEM with ITP approach for steady Schrödinger equation. International Journal of Computer Mathematics, 95:187-201, 2018.
[8] Luo Y., Gao S., Xie D., Dai Y. A discrete plant disease model with roguing and replanting. Advances in Difference Equations, 1:1-18, 2015.
[9] Chan M.S., Jeger M.J. An analytical model of plant virus disease dynamics with roguing and replanting. Journal of Applied Ecology, 31:413-427, 1994.
[10] Blas N., David G. Dynamical roguing model for controlling the spread of tungro virus via Nephotettix Virescens in a rice field. Journal of Physics: Conference Series, 893, 012018, 2017.
[11] Anggriani N., Yusuf M., Supriatna A.K. The effect of insecticide on the vector of rice tungro disease: insight from a mathematical model. International Information Institute (Tokyo), 20:6197-206, 2017.
[12] Anggriani N., Istifadah N., Hanifah M., Supriatna A.K. A mathematical model of protectant and curative fungicide application and its stability analysis. IOP Conference Series: Earth and Environmental Science, 31, 012014, 2016.
[13] Anggriani N., et al. Optimal control of plant disease model with roguing, replanting, curative, and preventive treatment. In Journal of Physics: Conference Series, 1657, 012050, 2020.
[14] Anggriani N., et al. Dynamical analysis of plant disease model with roguing, replanting and preventive treatment. In Proceedings of 4th International Conferenve on Research, Implementation, and Education of Mathematics and Science, Yogyakarta, 14–16 May 2017.
[15] Odibat Z., Kumar S. A robust computational algorithm of homotopy asymptotic method for solving systems of fractional differential equations. Journal of Computational and Nonlinear Dynamics, 14, 081004, 2019.
[16] Momani S., Abu Arqub O., Maayah B. Piecewise optimal fractional reproducing kernel solution and convergence analysis for the Atangana–Baleanu–Caputo model of the Lienard’s equation. Fractals, 28, 2040007, 2020.
[17] Akgul A., Cordero A., Torregrosa J.R. Solutions of fractional gas dynamics equation by a new technique. Mathematical Methods in the Applied Sciences, 43:1349-1358, 2020.
[18] Singh J., Swroop R., Kumar D. A computational approach for fractional convection-diffusion equation via integral transforms. Ain Shams Engineering Journal, 9:1019-1028, 2018.
[19] Caputo M., Fabrizio M. A new definition of fractional derivative without singular kernel. Progress in Fractional Differentiation and Applications, 1:73-85, 2015.
[20] Abu Arqub O., Singh J., Maayah B., Alhodaly M. Reproducing kernel approach for numerical solutions of fuzzy fractional initial value problems under the Mittag–Leffler kernel differential operator. Mathematical Methods in the Applied Sciences, 46(7):7965-7986, 2023.
[21] Baleanu D., Mousalou A., Rezapour S. On the existence of solutions for some infinite coefficient-symmetric Caputo-Fabrizio fractional integro-differential equations. Boundary Value Problems, 1:1-9, 2017.
[22] Teodoro G., et al. A review of definitions of fractional derivatives and other operators. Journal of Computational Physics, 388:195-208, 2019.
[23] Atangana A., Baleanu D. New fractional derivatives with nonlocal and non-singular kernel: theory and application to heat transfer model. Thermal Science, 20:763-769, 2016.
[24] Maayah B., Abu Arqub O., Alnabulsi S., Alsulami H. Numerical solutions and geometric attractors of a fractional model of the cancer-immune based on the Atangana-Baleanu-Caputo derivative and the reproducing kernel scheme. Chinese Journal of Physics, 80:463-483, 2022.
[25] Abu Arqub O., Singh J., Alhodaly M. Adaptation of kernel functions‐based approach with Atangana–Baleanu–Caputo distributed order derivative for solutions of fuzzy fractional Volterra and Fredholm integrodifferential equations. Mathematical Methods in the Applied Sciences, 46:7807-7834, 2023.
[26] Djennadi S., Shawagfeh N., Abu Arqub O. Well-posedness of the inverse problem of time fractional heat equation in the sense of the Atangana-Baleanu fractional approach. Alexandria Engineering Journal, 59:2261-2268, 2020.
[27] Akgül A. A novel method for a fractional derivative with nonlocal and non-singular kernel. Chaos, Solitons Fractals, 114:478-482, 2018.
[28] Jajarmi A., Ghanbari B., Baleanu D. A new and efficient numerical method for the fractional modelling and optimal control of diabetes and tuberculosis co-existence. Chaos: An Interdisciplinary Journal of Nonlinear Science 29, 093111, 2019.
[29] Yildiz T.A., et al. New aspects of time fractional optimal control problems within operators with nonsingular kernel. Discrete & Continuous Dynamical Systems-S, 13:407-28, 2020.
[30] Sardar T., et al. A generic model for a single strain mosquito-transmitted disease with memory on the host and the vector. Mathematical Biosciences, 263:18-36, 2015.
[31] Podlubny I. Fractional differential equations. Academic Press, New York (1999).
[32] Gorenflo R., Mainardi F. Fractional calculus: Int and differential equations of fractional order. In A. Carpinteri, F. Mainardi (Eds.), Fractals and Fractional Calculus, New York, 1997.
[33] Driessche P., Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathematical Biosciences, 180:29-48, 2002.
[34] Castillo-Chavez C., Feng Z., Huang W. On the computation of R0 and its role in global stability. IMA Volumes in Mathematics and Its Applications, 125:229-250, 2002.
[35] Diekmann O., Heesterbeek J.A.P. Mathematical epidemiology of infectious diseases: Model building, analysis and interpretation. John Wiley & Sons, Vol. 5, pp. 320, 2000.
[36] Nise N.S. Control systems engineering. John Wiley & Sons, 8th edition, pp. 800, 2020.
[37] Verhulst F. Nonlinear differential equations and dynamical systems. Springer, Heidelberg, pp. 306, 1996.
[38] Wiggins S. Introduction to applied nonlinear dynamical systems and chaos. Part of the book series: Texts in Applied Mathematics (TAM, volume 2), 2nd Edition, Springer-Verlag, Heidelberg, pp. 882, 2003.
[39] Kumar D., Singh J., Baleanu D. Analysis of regularized long-wave equation associated with a new fractional operator with Mittag-Leffler type kernel. Physica A, 492:155-167, 2018.
[40] Diethelm K., Freed A.D. The FracPECE subroutine for the numerical solution of differential equations of fractional order. Forschung und wissenschaftliches Rechnen, 57-71, 1999.
[41] Garrappa Z.R. On linear stability of predictor-corrector algorithms for fractional differential equations. International Journal of Computer Mathematics, 87:2281-2290, 2010.
[42] Butcher J. Numerical methods for ordinary differential equations in the 20th century. Journal of Computational and Applied Mathematics, 125:1-29, 2000.Document information
Published on 21/07/23
Accepted on 16/07/23
Submitted on 25/01/23
Volume 39, Issue 3, 2023
DOI: 10.23967/j.rimni.2023.07.001
Licence: CC BY-NC-SA license
Share this document
Keywords
claim authorship
Are you one of the authors of this document?