The dentate gyrus is a unique part of the brain because it is known for housing neurogenesis in the adult brain, a process which normally stops early in development. This makes the dentate gyrus an area of great interest, especially in combating neurodegenerative diseases. This area, as well as the entire nervous system, is composed of both neurons, which send and receive signals, and glial cells, which are responsible for supporting neurons. Glial cells including astrocytes, oligodendrocytes, and ependymal cells are present in both the central nervous system and peripheral nervous system, while Schwann Cells are present only in the peripheral nervous system, and microglia are present only in the central nervous system. Apoptosis and autophagy are both processes which degrade and recycle materials. Autophagy degrades materials inside the cell, such as organelles and proteins, and uses lysosomes to carry out the process. Apoptosis consists of degrading old or damaged cells, and is known as programmed cell death. Many proteins affect these processes including Beclin-1, UC3, p62, and the BCL-2 family of proteins. Some studies have used machine learning algorithms in conjunction with simulation and statistical software to study these proteins and the processes they are involved in. Several areas of research remain unfilled, especially in the area of glial cells and the proteins involved in autophagy and apoptosis, as not many machine learning studies have examined this.
1. Introduction
This review paper looks at the nervous system, including neurons and 5 different glial cell types and how they contribute to the larger nervous system. Form, function, and how they relate to different neurological diseases are looked at for astrocytes, microglia, Schwann Cells, oligodendrocytes, and ependymal cells. The processes of autophagy and apoptosis are also examined, and how certain proteins play a role in these processes is discussed. Proteins discussed include Beclin-1, LC3, p62, and the BCL-2 family of proteins. Previous experiments which used machine learning algorithms, alongside simulation and statistical software, to study these proteins are also examined, and the machine learning algorithms, datasets, and other software is identified.
The nervous system is split into 2 parts: the central and peripheral nervous systems. These are both made up of 2 different types of cells: neurons (see Figure 1), which send and receive signals, and glial cells, which support the neurons. Although most glial cells including astrocytes (see Figure 2), microglia (see Figure 3), oligodendrocytes (see Figure 5), and ependymal cells (see Figure 6) are present only in the central nervous system, some glial cells, like Schwann Cells (see Figure 4), can only be found in the peripheral nervous system, while neurons are found in both. Glial cells, although all meant to fulfill supportive roles, all differ in structure and function, and dysfunction of glial cells can be caused by or led to neurological diseases.
Autophagy and apoptosis are both processes that involve the cleansing of the nervous system. Autophagy refers to degradation of subcellular components such as damaged or old organelles, while apoptosis, or programmed cell death, refers to the degradation of entire cells that are damaged or old. Both processes recycle organic matter to be used to build new cells, organelles, and proteins.
Few studies exist that have used machine learning to study proteins such as Beclin-1, LC3, p62, or the BCL-2 family of proteins. Among these, machine learning algorithms such as K-nearest neighbor, logistic regression, gradient boosting, support vector value, and random forest are used, sometimes in congruence with QSAR. Supportive software that can be either simulation or statistical is commonly used, especially AMBER 14.0 and FF14SB. Software such as Smina, Plants, R software, Cytoscpae, Cox regression analysis, and others are also used.
2. Machine Learning
Machine learning has been used alongside in vivo and in vitro experiments in order to study things such as the BCL-2 family members, Beclin-1, and prognostic proteins of cancers. Machine learning algorithms such as K-nearest neighbors, logistic regression, gradient boosting, support vector machine, and random forest are imputed with both publicly available datasets and clinical datasets, and sometimes used with QSAR, in order to study these proteins. In these types of studies it is common for other simulation and statistical software to be used such as AMBER, FF14SB force field, Smina, Plants, Cox regression analysis, Cytoscape, MCODE, SHAKE algorithms, and Search Tool for the Retrieval of Interacting Genes.
A study that aimed to identify new inhibitors of anti-apoptotic members of the BCL-2 family used both in vivo and in vitro experiments, and a virtual screening protocol to find 2 potential new lead compounds, IS20 and IS21, both of which could bind into the Beclin-1 BH3 binding site of wild type BCL-2, BCL-XL, and Mcl-1, as well as reduce tumor sphere formation, cell viability, clonogen ability, and induce apoptosis in leukemic, melanoma and lung cancer cells (Valentini et al 2022). Using a ligand-based VS protocol set up to select potential BCL-2 modulator compounds from about 1 million compounds on a commercial database, it began with a compilation of a BCL-2 modulators dataset interrogating publicly available (Valentini et al 2022). ChEMBL and PubChem databases and retrieving all records related to ligands assayed against BCl-2 proteins (Valentini et al 2022). In order to use QSAR models, the data from ChEMBL was pruned and split into 2 groups: the binary assay set and the functional assay set (Valentini et al 2022). By using Python version 3.5 coding environment using anaconda, QSAR models were built (Valentini et al 2022). At first QSAR models were constructed by describing molecules in chemical representation using molecular descriptors, and molecular representation using molecular fingerprints. Several binary classification models were built with machine learning algorithms K-nearest neighbors, logistic regression, gradient boosting, support vector machine, and random forest (Valentini et al 2022). 42 total classification models were derived (Valentini et al 2022). The best QSAR models then ranked molecules from a commercial vendor called VITAS-M based on their promise as potential BCL2M (Valentini et al 2022). Molecular docking simulations were then carried out by free academia programs Smina and Plants. Imputed with the structures of 1 wild type BCL-2 protein and 3 mutated BCL-2 protein, UCSF Chimera was used, and the structures were aligned by means of a matchmaker module (Valentini et al 2022). First the complexes were geometrically optimized with a short single point minimization with embedded AMBER minimization core and FF14SB force field with 1000 steepest descent steps and 100 conjugate gradient steps (Valentini et al 2022).
Another study used mainly computation methods to test viral BCL-2 proteins and the binding-associated conformational changes and presence of long-range interactions within these viral BCL-2 proteins (Ramanathan et al 2020). This study used molecular dynamics simulations for both the apo- and holo- forms of BCL-2 (Ramathan et al). 3 structures were used: apo form of BCL-2 from the NMR ensemble, holo form of BCL-2 in complex with Beclin-1, and apo-M11 initialized without Beclin-1 bound to the binding site (Ramanathan et al 2020). Each of the 3 systems was processed using the AMBER 14 suite of molecular modeling software, with the FF14SB force-field (Ramanathan et al 2020). Production runs were performed using AMBER 14.0, with parameters of a constant number of particles, constant volume, energy (NVE) ensemble, with particle mesh Ewald technique for electrostatics and a 10 amps cut-off for Lennard-Jones interactions (Ramanathan et al 2020). Motion of all covalently bound hydrogen atoms was restricted using the SHAKE algorithm (Ramanathan et al 2020). Simulations were performed at a pressure of 1 atm and temperature of 300 K, and data from simulation were stored every .05 ns (Ramanathan et al 2020). A computation analysis of M11 revealed an extensive network of hydrophobic interactions (Ramanathan et al 2020). A TD4 module was designed that performed joint diagonalization of time-delayed cumulant matrices, and was the counterpart of SD4, using the assumption that a molecular simulation trajectory is a linear combination of independent, anharmonically fluctuating protein motions (Ramanathan et al 2020). Blind Source Separation was used to discover these anharmonic motions (Ramanathan et al 2020).
Another study used computational biology to identify potential prognostic biomarkers of bladder cancer (Xu et al 2021). A large-scale multi-comic dataset from the Cancer Proteome Atlas was used and immune infiltration, survival and other statistical analyses were implemented using R software in cancers, where n = 12,452 (Xu et al 2021). In the end, a landscape of prognostic protein signatures of urinary bladder cancer patients was identified (Xu et al 2021). It was found that elevated SRC expression contributes to an immunosuppressive microenvironment mediated by immune checkpoint molecules of urinary bladder cancer (Xu et al 2021). SRC was then validated in vivo as a potential biomarker in predicting response to immune checkpoint therapy (Xu et al 2021). It was found that expression of SRC in the baseline tumor tissues correlates with improved survival benefits, but predicts worse immune checkpoint therapy response (Xu et al 2021). Clinical data and gene expression profiles for patients with urinary bladder cancer were obtained from the Cancer Genome Atlas, and the level 4 data of reverse-phase protein arrays were obtained from the Cancer Proteome Atlas (Xu et al 2021). R software was used to obtain a total of 341 urinary bladder cancer cases with full biological microarray and clinical data in order to impute the missing data and match the sample ID (Xu et al 2021). Prognostic values of proteins were assessed using the Kaplan-Meier method and the hazard ratio estimates with univariate Cox regression analysis used to estimate 95% confidence intervals (Xu et al 2021). Lasso Cox regression analysis was then performed to further restrict the candidate proteins (Xu et al 2021). Risk scores of patients were calculated with multivariate analysis with expression of proteins and survival rates (Xu et al 2021). Patients were then divided into low or high risk levels, and survival curves and scatter diagrams of patient prognosis and risk score were plotted using R software (Xu et al 2021). An integrated prognosis-related proteins model was constructed by univariate and multivariate Cox regression analyses (Xu et al 2021). New patient data was drawn from Cancer Genome Atlas and Gene Expression Omnibus, and single-cell RNA sequencing datasets from Tumour Immune Single-cell Hub were inputted (Xu et al 2021). Both Pearson correlation analysis and Kaplan-Meier method were utilized. Gene Set Enrichment Analysis then used the Cancer Genome Atlas datasets, and protein-protein interaction networks for differential expressed genes was predicted using Search Tool for the Retrieval of Interacting Genes version 10.0 (Xu et al 2021). Cytoscape version 3.5 and MCODE version 1.4.2 were used to visualize molecular interaction networks of differential expressed genes (Xu et al 2021).
3. Dentate Gyrus
Located in the mammalian hippocampus, the dentate gyrus is responsible for memory, emotion, and spatial memory, and its neurons play a key role in transmitting pain-related information. It is unique because it is the only part of the brain where adult neurogenesis occurs. This makes it an area of interest when studying neurodegenerative diseases such as Alzheimer’s Disease, as adult neurogenesis may be able to negate neurodegeneration.
3.1. Composition and purpose
The dentate gyrus is one of three main parts in the mammalian hippocampus, along with the subiculum and the hippocampus proper (Ma et al 2024). The hippocampus itself plays an important role in memory, emotion and spatial memory (Ma et al 2024). Three distinct layers, the molecular layer, the granule cell layer, and the polymorphic layer, make up the dentate gyrus (Shahid 2023). The molecular layer contains the dendrites of the dentate granule cells, while the granule cell layer is made of mainly granule cells closely packed together, and the polymorphic layer is composed mostly of mossy cells and lies within the granule layer (Shahid 2023). The major cell type in the dentate gyrus is granule cells, originating from the neural progenitor cells in the dentate neuroepithelium or primary matrix (Ma et al 2024). It has been determined that neurons in the dentate gyrus play an essential role in processing and modulating pain-related information, and many neurons in the dentate gyrus are solely associated with pain pathways as they respond to painful stimuli (Nazari-Serenjeh et al 2024).
3.2. Neurogenesis
The adult dentate gyrus is unique because just as the peripheral nervous system of adult mammals are capable of neuronal regeneration, the dentate gyrus can produce new neurons throughout life (Liu et al 2024). This intrinsic process includes neural stem cells, or neural precursor cells, driving the production of new neurons in the substratum of the dentate gyrus (Liu et al 2024). These resident neural stem cells produce new granule cells which are adopted into the larger hippocampal network as they receive synapses from cortical neurons from the perforant path, and then form synapses with downstream neurons (Dieni et al 2019). Neuronal regeneration is regulated by various transcription and growth factors and affected by several biomarkers such as doublecortin, neuronal nuclear antigens, proliferating cell nuclear antigen, and APP (Liu et al 2024). Despite a majority of these newly created neurons undergoing programmed cell death, the surviving neurons are incorporated permanently into the dentate gyrus and acquire properties similar to the granule cells created earlier in the life cycle, in addition to increasing the granule cell population of the dentate gyrus (Diene et al). Neurogenesis in the adult dentate gyrus seems to be regulated by extrinsic factors such as environmental enrichment and physical exercise (Bihelek 2019). It is known that an increase in exercise such as running can improve the rate of neurogenesis in the adult dentate gyrus, as it increases the rate of proliferation of neural progenitor cells, and accelerates the maturation and functional integration of developing neurons (Trinchero et al 2019). Environmental enrichment also increases survival and accelerates maturation of new granule cells, despite having no influence on proliferation (Trinchero et al 2019).
3.3. Combating Alzheimer’s Disease
Neurogenesis in the dentate gyrus is a process that may have the capacity to combat neurodegenerative diseases such as Alzheimer’s Disease. Although there are no existing medications that can cure Alzheimer’s Disease, and only a few symptoms can be treated by drugs, it has been shown that the pathological process underlying neurodegenerative diseases such as Alzheimer’s Disease and cognitive impairment can be fought by improving the dysfunction of neurogenesis with alternative forms of medicine such as moxibustion and acupuncture (Liu et al 2024). In addition it has been shown that the induction of extensive transcriptional and epigenetic changes in damaged neurons and the promotion of the proliferation and differentiation of adult hippocampal neural progenitor cells through the Shh signaling pathway have both shown promise in fighting Alzheimer’s Disease in patients (Liu et al 2024).
4. Nervous system cell population
The nervous system, responsible for receiving, transmitting, analyzing and acting upon information, is broken up into 2 main parts: the peripheral nervous system and the central nervous system. The peripheral nervous system is responsible for receiving information from the outside world and transmitting it to the brain, and then taking action based upon what the brain decides to do. The central nervous system, made up of the brain and spinal cord, is responsible for analyzing and deciding what to do based upon information. The nervous system is composed of 2 major cell types: neurons and glial cells, which include astrocytes, microglia, schwann cells, oligodendrocytes, and ependymal cells.
4.1. Neurons
4.1.1. Peripheral nervous system
Neurons (see Figure 1) are one of the major cell types and are the core of both the central nervous system and the peripheral nervous system. The peripheral nervous system includes sensory neurons, which carry information from the sensory receptor cells, motor neurons, which transmit information from the brain to the muscles and glands, causing them to take action, and interneurons, which carry the signal and connect the sensory neurons to the motor neurons (Cherry 2023).
4.1.2. Central nervous system
The central nervous system consists of both the brain and the spinal cord, both of which are protected by bone, as the brain is protected by the skull and the spinal cord is protected by the vertebrae (Hirsch et al 2022). Both are also cushioned by membranes and cerebrospinal fluid, which flows through the ventricles of the brain and in the spinal column (Hirsch et al 2022). Aside from cushioning, cerebrospinal fluid also takes away waste products and nourishes the central nervous system (Hirsch et al 2022).
The brain, made up of the cerebrum, the brainstem, and the cerebellum, is responsible for controlling all of the body's functions, even down to the ones we are less aware of such as a heart beating and digestion (Hirsch et al 2022). The spinal cord allows the brain to communicate with the peripheral nervous system, which sends signals to the rest of the body (Hirsch et al 2022).
4.1.3. Structure
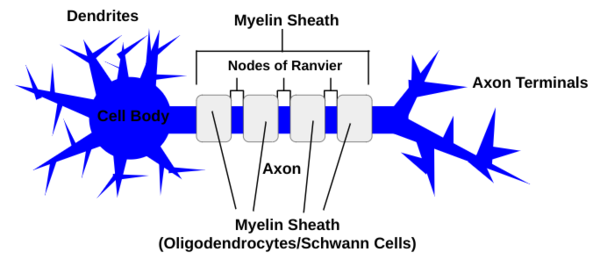
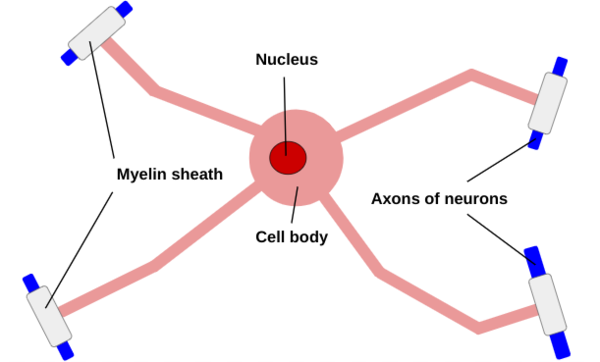
The three basic parts of the neuron include the cell body, which produces proteins, the dendrites, which extend off of the cell body and receive chemical signals from other neurons via one of the thousands of connections a single neuron in the brain can create, and the axon, which also extends from the cell body and carries the electrical signal to other neurons (see Figure 1) (Cherry 2023). The axons are insulated in the myelin sheath, which is composed of oligodendrocytes in the central nervous system, and schwann cells in the peripheral nervous system (Wei et al 2024). Axon diameters correlate with myelin sheath length (Bechler et al 2015).
Figure 1: A neuron is composed of dendrites, which receive chemical signals from other neurons and extend from the cell body, an axon that transports an electrical signal and is insulated by the myelin sheath, which is made up of either oligodendrocytes or Schwann Cells separated by nodes of Ranvier, and axon terminals which send chemical signals to other neurons.
4.1.4. Signaling
First the receptor sites on the dendrites of the neuron receive a signal from neurotransmitters that either stimulate activity, triggering depolarization in the cell body towards the axon hillock, or dampen activity, making it harder for a neuron to fire (Cherry 2023). Depolarization is characterized by sodium ions entering the neuron, changing the electrical charge from its base of -70 millivolts (Cherry 2022). The signal will continue at the axon hillock only if the change in electric charge is great enough according to the all-or-none law, usually -55 millivolts, and then an action potential sends an electrical signal down the axon to the next synapse (Cherry 2022). Myelin sheath length impacts axonal conduction velocity because it influences the spacing between the nodes of Ranvier, the open space in the myelin sheath, affecting neural signal coordination and synchronization (Bechler et al 2015). It promotes rapid and efficient nerve conduction (Salzer 2105). Many but not all synaptic gaps require neurotransmitters to be released by the presynaptic neuron such as acetylcholine, which is associate with memory, learning, muscles contractions, and Alzheimer’s Disease when not sufficient, endorphins, which are associated with pain perception, sexual response and maternal behavior, dopamine, which is associate with pleasurable feelings, Parkinson’s Disease when not sufficient, and schizophrenia when in excess, and serotonin, which is associated with mood stabilization, learning, memory, blood clotting, digestion, bone health and sleep (Cherry 2023). To stop the signal between neurons and to prepare for the signal to be sent again, either the presynaptic neuron reabsorbs the neurotransmitters via reuptake, the neurotransmitters are broken down in the synaptic gap by enzymes via degradation, or the neurotransmitters diffuse and reset (Cherry 2023).
4.2. Glial cells
Glial cells are cells that are part of the nervous system but do not participate directly in synaptic interaction and electrical signaling, and instead perform supportive functions that help define synaptic contacts and maintain the signaling abilities of neurons (Purves et al 2001). They are smaller than neurons and without axons and dendrites (Purves et al 2001). Glial cells make up about half of the central nervous system, although it was previously believed that they greatly outnumber neurons, influence and aid in neuronal birth, migration, axon specification and growth, and even circuit assembly and synaptogenesis (Allen et al 2018).
4.2.1. Astrocytes
The most abundant glial cells in the mammalian brain, astrocytes (see Figure 2) are crucial for the proper health and function of the nervous system, as they provide important metabolic and trophic support to neurons, and astrocyte formation usually occurs after the main periods of neurogenesis and neuronal migration are completed, except in the dentate gyrus, where astrocytes provide signals that participate in the generation of new neurons and their integration into existing circuits (Allen et al 2017). Astrocytes are located only in the central nervous system (Rasband 2016). Astrocytes perform metabolic, structural, homeostatic and neuroprotective tasks such as clearing excess neurotransmitter, stabilizing and regulating the blood-brain barrier, and promoting synapse formation, all without conducting electrical signals, but instead ensuring the continued function of neurons (Wei et al 2024). Astrocytes are given their name due to their radially-arranged foot processes, giving them a star-like appearance and are marked by glial fibrillary acidic protein, which allows astrocytes to communicate with and affect surrounding neural vasculature (Wei et al 2024). Astrocyte-neuron interactions are critical for the proper functioning of synapses and neural circuits, as astrocytes express factors involved in triggering synapse formation (Ngoc et al 2024). Across many species, astrocytes secrete numerous proteins that regulate distinct facts of synapse formation and transmission, and astrocytes mediate bidirectional communication with both the presynaptic and postsynaptic neuron due to their proximity to the synaptic cleft through their PAPs (Ngoc et al 2024). Using both exocytotic secretion and adhesion, astrocytes signal directly to neuronal synapses (Ngoc et al 2024). Coupling at gap junctions, astrocytes form a network that coordinates their responses to stimuli such as stress, and are broken into four main types: interlaminar, protoplasmic, varicose projection, and fibrous (Wei et al 2023).
Figure 2: Astrocytes get their name because of their radially-arranged foot processes that give them a star-like appearance. These processes branch out from the cell body, which is home to the nucleus. Astrocytes exist solely in the central nervous system.
Unique to humans and primates, interlaminar astrocytes have both radial tangential fibers and vertical processes, and get their name because these astrocytes project their vertical processes, which can be up to one millimeter in length, to layers III and IV of the cerebral cortex, despite interlaminar astrocytes originating in layer I (Wei et al 2023). These astrocytes have small cell bodies, have a high density of GFAP fibers, and are in abundance within the human central nervous system (Wei et al 2023).
The majority of astrocytes are protoplasmic, which are large with a high density of tortuous, bulbous GFAP-positive processes then branch into smaller and finer processes (Wei et al 2023). These astrocytes are responsible for modulation and synchronization of blood flow with synaptic activity to improve neural efficiency (Weit et al 2023). They are distributed uniformly throughout the grey matter in cortical layers II, III, IV, V, and VI, and organize into separate domains in which each individual astrocyte, with little overlap, projects to the surrounding vasculature and synapses (Wei et al 2023).
Fibrous astroglia are located around white matter tracts and are larger than protoplasmic astrocytes, despite expressing fewer foot processes (Wei et al 2023). Due to white matter having few synapses, fibrous astroglia communicate almost exclusively with the neurovasculature to regulate metabolic and ionic homeostasis in the central nervous system instead of regulating synaptic activity (Wei et al 2023).
Occupying cortical layers V and VI, the varicose projection astroglia have long foot processes that may connect with cells in other cortical layers, just like interlaminar astrocytes, and may be up to one millimeter in length, and have spiny branches and varicosities every 10 micrometers (Wei et al 2023). They are thought to allow for higher-order cognitive processing, due to the fact that only primates contain this astroglial type, despite their function being unknown (Wei et al 2023).
4.2.2. Microglia
Microglia (see Figure 3) are found within the central nervous system only and are known as the resident macrophages of the brain parenchyma, and are responsible for carrying out diverse roles (Munro et al 2024). Although not completely understood, microglia may promote the maturation and maintenance of other brain cells such as macroglia and neuronal cells (Munro et al 2024). Microglia are located exclusively in the central nervous system, and prefer white matter, although they exist in gray matter (Bozidis 2024). Higher concentrations can be found in the brainstem, hippocampus, basal ganglia, and areas with extensive neuronal degeneration, as cell numbers increase in response to recent trauma or inflammation, actively encircling and penetrating affected tissues in order to perform their immune functions (Bozidis 2024). Microglia are the smallest type of glial cells, and can be distinguished due to their small and elongated nuclei, while other glial cells have larger and more rounded nuclei (Bozidis 2024). As a part of the mononuclear phagocyte system, microglia are macrophages, which can arise from 3 distinct developmental pathways (Prinz et al 2019). In the resting state, microglia boast numerous short and unevenly-shaped processes branching out in various directions, which constantly change in length and shape (Bozidis 2024). In the activated state, microglia initiate vesicle and protein production, and the branches shorten and thicken as the cell’s body increases in size and eventually adopts an amoeboid shape with almost no visible processes, and the cell membrane appears folded (Bozidis 2024). In the phagocytic state, the cells increase in size and accumulate lipid granules, as well as adopting a distinctive appearance known as a gitter cell, which is typically found in regions of the central nervous system experiencing infection, inflammation, or trauma (Bozidis 2024).
Figure 3: Microglia are composed of a cell body with an elongated nucleus, and several processes that branch out from the cell body. How big the cell body and the processes are depend on which phase the cell is in: resting state, activated state, or phagocytic state. Microglia exist solely in the central nervous system.
Although they act as the first line of defense in the brain by phagocytosing harmful pathogens and cellular debris, chronic microglia activation can lead to irreversible damage to the central nervous system (Muzio et al 2021). Microglia emerge from early erythromyeloid progenitors of the yolk sac and enter the developing brain before a fully mature blood-brain barrier is established (Muzio et al 2021). Microglia contribute to central nervous system homeostasis during brain development by supporting cell proliferation of neural precursors, and have the ability to regenerate shortly (Muzio et al 2021). After birth, microglia sculpt synapses, and after injury to the central nervous system, they change their morphology and donw-regulate genes supporting homeostatic functions (Muzio et al 2021). Microglia are highly dynamic and are considered among the most versatile cells in the body, as they can morphologically and functionally adapt to their environment, and are key components of central nervous system homeostasis, as microglial dysregulation can lead to neurological disease (Nayak et al 2014). The activation of both microglia and astrocytes is heterogeneous and categorized as either neurotoxic in M1-phenotype microglia nd A1-phenotype astrocytes, or neuroprotective in M2-phenotype microglia and A-2 phenotype astrocytes (Kwon et al 2020). In an experiment with mice, it was shown that non-microglial brain cells can reach their mature transcriptomic states despite an absence of microglia (Munro et al 2024). Microglial cells play a key role in depression and stress, but can be countered by quercetin, which inhibits the expression of STAT1 in microglia and the sympathetic nervous system activity in a chronic stress-induced tumor (Luo et al 2024). There is evidence to suggest that regulating microglia functions during disease pathology may be a strategy to counteract brain degeneration in multiple sclerosis, Alzheimer’s Disease, Parkinson’s Disease, and amyotrophic lateral sclerosis (Muzio et al 2021).
4.2.3. Schwann cells
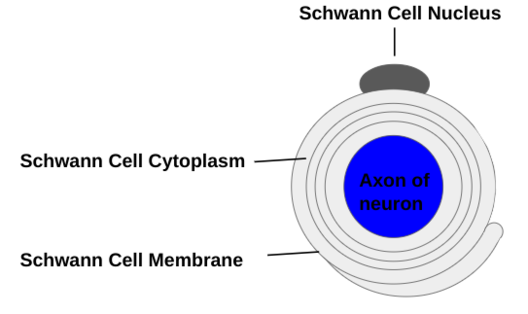
Schwann cells (see Figure 4) form the myelin sheath in the peripheral nervous system and are not found within the central nervous system, and they are superior in phagocytosis of myelin debris, making nerve regeneration superior in the peripheral nervous system (Wei et al 2024). Myelinating Schwann Cells are both radially and longitudinally polarized cells, and are organized with myelination into distinct membrane domains, each with a unique array of proteins, and a communicating set of cytoplasmic compartments (Salzer 2015). Although both Schwann Cells and oligodendrocytes produce myelin, there are key differences in the way it is developed and assembled in the 2 nervous systems (Salzer 2015). Peripheral nervous system axonal neuregulin-1 type III regulates the initiation and properties of Schwann cell myelin sheaths, something that is unique to the peripheral nervous system (Bechler et al 2015).
Figure 4: Schwann Cells, existing solely in the peripheral nervous system, surround the axons of neurons to form the myelin sheath. Schwann Cells have the nucleus near the top of the cell, and slowly grow around and insulate the neuronal axon in a radial pattern.
4.2.4. Oligodendrocytes
Oligodendrocytes (see Figure 5) make up the myelin sheath in the central nervous system, and respond to fiber diameters, have regional identity and generate different sheath lengths that mirror internodes, and self-regulate the formation of compact, multilamellar myelin and generate sheaths of physiological length (Bechler et al 2015). Oligodendrocytes do not exist in the peripheral nervous system (Rasband 2016). During developmental myelination of the central nervous system, oligodendrocyte progenitor cells proliferate and migrate, and then terminally differentiate into mature oligodendrocytes and myelinate axons, a highly regulated process in which extracellular and intrinsic signals regulate transcriptional and physiological changes in oligodendrocyte lineage cells (Elbaz et al 2019). Unlike schwann cells, they are not sufficient at nerve regeneration, making the central nervous system more vulnerable and susceptible to injury and disease (Wei et al 024). Making up about 5-10% of the total glial cell population, oligodendrocytes and their precursors are very susceptible to injury by excitotoxic damage, oxidative stress, inflammatory events and other methods (Barateiro et al 2016). Oligodendrocyte lineage cells include mature oligodendrocytes and oligodendrocyte progenitor cells, and may participate in the pathogenesis of depression (Zhou et al 2020).
Figure 5: Oligodendrocytes form the myelin sheath and exist in the central nervous system only. The nucleus is housed in the cell body, where several branches extend from to hold in place myelin on the axons of neurons, therefore insulating them.
4.2.5. Ependymal cells
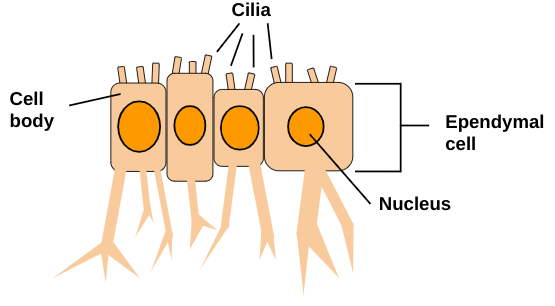
Ependymal cells (see Figure 6) are ciliated cells that line the brain ventricles and the spinal cord central canal and key components of the ventricular-subventricular zone and central canal stem cell niches (Albors et al 2023). They are located in the central nervous system only (Rasband 2016). Arising early in embryogenesis, ependymal cells are unique because of their cell heterogeneity and damage-responsive neural stem cell potential (Frederico et al 2022). It is known in the adult mouse brain the ependymal cells are postmitotic and the neural stem cell potential resides within subependymal adult neural stem cells, as ependymal cells can generate scar-forming astrocytes and oligodendrocytes following spinal cord injury, yet oligodendrocytes are not replaced sufficiently and new neurons are not generated, so function is not restored as it previously was (Albors et al 2023). In organisms such as axolotls, zebrafish and mice it has been shown that spinal cord ependymal cells can self-renew and generate specialized cells after injury, although the ability to do so varies greatly between species, as in humans ependymal cells in the spinal cord have not been observed proliferating, despite displaying neural stem cell characteristics when cultured (Albors et al 2023). Motile cilia are located on the surface of ependymal cells in all brain ventricles, and contribute to the flow of cerebrospinal fluid (Delgehyr et al 2015). Spinal cord ependymal cells have been found to display neural stem cell properties and generate scar-forming astrocytes and remyelinating oligodendrocytes after injury (Stenudd et al 2022).
Figure 6: Ependymal cells house their nuclei in the cell body. Cilia are located on the surface of the cell and contribute to the flow of cerebrospinal fluid. Ependymal cells line brain ventricles, the spinal cord central canal, key components of the ventricular-subventricular zone, and the central canal stem. Ependymal cells exist solely in the central nervous system.
5. Autophagy
Autophagy is part of the stress response and a lysosomal degradative pathway which regulates the homeostasis of eukaryotic cells, and can degrade misfolded or aggregated proteins, clear damaged organelles, and eliminate intracellular pathogens (Song et al 2024). As autophagy is a part of the immune response, certain viruses have evolved to be able to inhibit autophagy and evade the immune response, or even induce autophagy and then hijack autophagic components (Song et al 2024). Dysregulated mTOR signaling and autophagy allows for cancer cells to bypass quality control mechanisms and maintain their growth (Galhuber et al 2024). Autophagy is also known as an alternative pathway for the recycling and provision of metabolite intermediates as anabolic building blocks (Galhuber et al 2024). mTORC1 is known to inhibit the early stages of autophagy by phosphorylation of unc-51 like autophagy activating kinase 1 at S638 and S758, as unc-51 like autophagy activating kinase 1 phosphorylates the autophagy initiation complex and mediates its translocation to the isolation membrane and autophagosome formation (Galhuber et al 2024). A key process in autophagy is autophagosomes formation, which occurs in 3 main steps: initiation, where proteins of the unc-51 like autophagy activating kinase 1 complex combine at the phagophore assembly site, nucleation, where activated autophagy activated kinase complex targets a class III PI3K complex to contribute to the production of a PI3K pool specific to autophagosomes, and expansion, where phosphatidylinositol 3-phosphate-enriched membrane domains, or omegasomes, are formed and then expand to form double-membrane autophagosomes, and then the autophagosome membrane recruits the ATG12-ATG5-ATG16 complex which facilitates MAP1LC and LC3 lipidation with phosphatidylethanolamine (Mehrbod et al).
5.1. Macroautophagy
The most prevalent form of autophagy, macroautophagy, utilizes double-membrane vesicles called autophagosomes, and is usually referred to as autophagy (Song et al 2024). Macroautophagy degrades macromolecules such as proteins, carbohydrates and lipids, and mediates turnover of endosomes and organelles (Galhuber et al 2024). These double membrane structures, or isolation membranes, originate from the plasma membrane and intracellular compartments to form autophagosomes which isolate cytoplasmic macromolecules and organelles by selective or non-selective routes (Galhuber et al 2024). Autophagosomes fuse with lysosomes and form autolysosomes, and then are degraded by hydrolyzing enzymes into small molecules such as monosaccharides and amino acids that are transported to the cytoplasm for recycling (Song et al 2024).
5.2. Microautophagy
Microautophagy involves lytic organelles including endosomes or lysosomes taking up a portion of cytoplasm directly, is in a variety of organisms, and is one of the five ways in which degradative substrates get into the interior space, the lumen, of lysosomes or vacuoles (Kuchitsu et al 2023). In microautophagy, membrane proteins in the limiting membrane of early endosomes or late endosomes can be incorporated into intraluminal vesicles that bud from the limiting membrane, and during this process, the cytosolic components can also be incorporated into the intraluminal vesicles in either a nonselective or a selective manner, allowing lysosomal degradation of cytosolic components with endosomal microautophagy (Kuchitsu et al 2023). This process generates multivesicular bodies, which, through the fusion of the multivesicular bodies with lysosomes, membrane proteins in intraluminal vesicles reach the lysosomal lumen (Kuchitsu et al 2023).
5.3. Chaperone-mediated autophagy
Chaperone-mediated autophagy is another way in which degradative substrates get into the lumen of lysosomes or vesicles (Kuchitsu et al 2023). In this process a cytosolic chaperone protein, Hsc70c, recognizes KFERQ-like motifs present in the cytosolic proteins, and directs them to Lamp2A at lysosomal surface, followed by the translocation of the substrate proteins across the lysosomal membrane and into the lysosomal lumen (Kuchitsu et al 2023).
5.4. Proteins
5.4.1. Beclin-1
Beclin-1 is a protein that is encoded by humans by the Beclin1 gene, an essential autophagy gene (Li et al 2020). It was first discovered in yeast as ATG6/VPS30 (Tran et al 2021). As heterozygous knockdown Beclin1 is a defective protein of autophagy, it promotes the malignant transformation and spontaneous tumors (Li et al 2020). Beclin1 is a haploinsufficient tumor suppressor, and it is monoallelically deleted in 40-75% of both sporadic breast and ovarian cancers (Tran et al 2021). It plays a role in many processes including murine embryo development, dauer development, immunity, neuronal and cardiac health, worm intestinal function and embryo development, murine neuronal health, skin development, intestinal barrier function, and is essential in Drosophila for wing and blood development and salivary secretion (Tran el al 2021). Specifically, Beclin1 regulates endocytic trafficking and LC3-associated phagocytosis in addition to its role in autophagy (Tran et al 2021). Beclin-1 has been found to be reduced in a brain with Alzheimer’s Disease (Salminen et al 2013).
Beclin-1 has the ability to self-associate, due to it being a BH3-only domain protein (Kang et al 2011). Oligomerized Beclin-1 serves as a platform enabling rapid nucleation of its associating proteins (Kang et al 2011). The Bcl-2-binding domain of Beclin-1 is not essential for its self-interaction (Kang et al 2011).
5.4.1.1. Role in autophagy
As a regulator of autophagy, Beclin-1 functions in conjunction with other proteins to form Class III Phosphoinositide 3-Kinase complexes in order to generate phosphorylated phosphatidylinositol, which is a lipid essential for many membrane trafficking processes in addition to autophagy (Tran et al 2021). This structure is one of the most important protein complexes in the autophagy pathway, and acts from the formation of autophagosomes to their extension and maturation (Li et al 2021). A central regulator in mammalian cells, Beclin1 interacts with a multitude of proteins including ATG14L, UVRAG, Rucion, and Bcl-2. It has three functional domains including a N-terminal Bcl-2 homology 3 domain, which interacts with the Bcl-2 family protein Bcl-XL, a central coiled-coil domain, which mediates interaction of Beclin1 with ATG14L and UVRAG, and a C-terminal evolutionarily conserved domain, which mediates the interaction of Beclin1 with VPS34 and activation of VPS34 kinase activity to regulate the size and number of autophagosomes (Li et al 2020). These proteins, with Beclin-1, assemble into two different Class III PI3K complexes, Complex 1 and Complex 2, depending on whether ATG14 or UVRAG is present (Tran et al 2021). The catalytic lipid kinase subunit within these complexes, VSP34, is responsible for the phosphorylation of phosphorylated phosphatidylinositol, which then mediates autophagy and other membrane trafficking functions through recruitment of effector proteins (Tran el al 2021). It has been found that autophagy can be sustained, even under conditions of severe sepsis, by enhancing Beclin-1 signaling, which suppresses mTOR activation (Sun et al 2018).
5.4.2. LC3
All steps of autophagy are tightly regulated by evolutionarily conserved autophagy-related proteins, such as ATG8, which is the key autophagic ubiquitin-like protein involved in the formation of autophagophores, of which there are 3 subfamilies: microtubule-associated protein light chain 3 (LC3), Golgi-associated ATPase enhancer of 16 kDa, and gamma-aminobutyric acid receptor-associated protein (Hwang et al 2022). In humans, there are 3 LC3s (LC3A, LC3B, LC3C), 2 gamma-aminobutyric acid receptor-associated proteins (GABARAP and GABARAPL1), and one Golgi-associated ATPase enhancer of 16 kDa (GATE-16 or GABARAPL2) (Fan et al 2021). The LC3 mammalian isoforms of LC3A, LC3B, and LC3C, are subject to a posttranslational ubiquitin-like conjugation pathway, by which they are covalently conjugated to phosphatidylethanolamine and localized to the autophagosomal membrane (Hwang et al 2022). This is carried out by ATG7 activating LC3-I, which is then transferred to ATG3, which is an E2-like enzyme, and LC3-I is modified to its membrane-bound form of LC3-II (Tanida et al 2004). The conversion of LC3 from the unconjugated form of LC3-I to a phosphatidylethanolamine-conjugated form of LC3-II is key for the formation of autophagosomes (Hwang et al 2022). As autophagosomes fuse with lysosomes to create autolysosomes and intra-autophagosomal components are degraded by lysosomal hydrolases, LC3-II proteins in auto lysosomal lumen is degraded (Tanida et al 2008). As an autophagosomal marker in mammals, LC3-II has been used to study autophagy in neurodegenerative and neuromuscular diseases, tumorigenesis, and bacterial and viral infections (Tanida et al 2004). Specifically, confocal imaging of LC3 and other proteins such as p62, as well as western blot is used (Beccari et al 2023). LC3B itself is a central protein in autophagy, and direct binding of LC3B to mRNAs elicits rapid degradation of target mRNAs (Hwang et al 2022). Specifically, when LC3B binds to and rapidly decays PRMT1 mRNA, efficient autophagy is promoted (Hwang et al 2022). Autophagy can be inhibited by attenuating LC3B lipidation with DC-LC3, which reduces the extent of autophagic structure formation and later substrate degradation (Fan et al 2021).
Japanese encephalitis virus may use autophagy as an immune evasion mechanism, and infection can induce accumulation of LC3-II in neuronal cells, enhancing autophagosomes and autolysosomes in the brain (Ashraf et al 2021). In contrast, a study found a progressive decrease in LC3-associated phagocytosis in the striatal astrocytes of mice with Huntington's Disease (Wakida et al 2022). In the nervous system microglial debris is engulfed by astrocytes that use LC3-associated phagocytosis, a form of noncanonical autophagy, and the debris-containing LC3-associated phagosomes are fused with lysosomes in order to form phagolysosomes for further degradation (Zhou et al 2022). It has been found that autophagy occurs mainly in neurons, but not in astrocytes or microglia in the dentate gyrus of corticosterone mice (Zhange et al 2023).
5.4.3. p62
p62 is a multifunctional adaptor protein involved in several biological processes including cell signaling, differentiation and removal of toxic protein aggregates, known as selective autophagy (Shaid et al 2013). Selective autophagy consists of cytoplasmic components being selected and tagged, commonly with ubiquitin, before being sequestered into an autophagosome by selective autophagy receptors such as p62/SQSTM1 (Lamark et al 2017), also known as sequestosome 1 (Galluzzi et al 2019). This is because p62 possesses a ubiquitin-binding domain that allows the protein to be recognized and recruited onto structures marked with ubiquitin (Filali-Mouncef et al). Selective autophagy is regulated by the ability of p62 to co-aggregate with ubiquitinated substrates being strongly induced by post-translational modifications (Lamark et al 2017). p62 is known to work with a number of proteins including ATG16L1, ATG7, LC3, and CALCOCO2 to encapsulate cytoplasmic Toxoplasma gondii, a eukaryotic parasite, in multi-membrane vesicles that efficiently blunt parasite replication, yet do not fuse with endosomes or lysosomes for removal (Galluzzi et al 2019). Lack of autophagy can lead to an accumulation of p62 protein, which can induce cellular stress and lead to disease, particularly in liver cells (Rusten et al 2010). p62 levels can be influenced by ATG5, as expression of ATG5 in ATG5-deficient macrophages restores expression of the AG5-ATG12 conjugate and reduces p62 levels, while expression of ATG5/K130R generates an unconjugated form of ATG5 and increases p62 levels (Hwang et al 2012). In the nervous system, astroglia display increased levels of p62 protein (Szebényi et al 2021).
6. Apoptosis
As autophagy is a way to recycle damaged cellular organelles and protein aggregates, apoptosis is a way to recycle damaged or aged cells (Liu et al 2019). Balance between these 2 processes is crucial, especially in the long-living cells found in the brain and nervous system, as an imbalance is associated with neurodegenerative diseases such as Parkinson’s Disease (Liu et al 2019). B cell lymphoma-2 (BCL-2) family member proteins are crucial because they are involved in both apoptosis and autophagy (Liu et al 2019). Excessive apoptosis can promote myocardial ischemia, post-ischemia cardiac remodeling and coronary atherosclerosis, while activated autophagy can protect against these conditions (Dong et al 2019).
6.1. BCL-2 family proteins
BCL-2 family proteins play a crucial role in apoptosis (Kale et al 2018), and block autophagy (Pattingre et al 2006). The BCL-2 family of proteins is responsible for initiating apoptosis by direct binding interactions that regulate mitochondrial outer membrane permeabilization, which leads to the release of intermembrane space proteins, and then caspase activation and apoptosis (Kale et al 2018). Mitochondrial outer membrane permeabilization is a rapid and complete process defined by cytochrome c being released from most mitochondria within 5 minutes and complete caspase activation occurring within 15 minutes, which then leads to apoptosis (Kale et al 2018). It has been found that overexpression of yes-associated proteins could increase BCL-2 protein level and suppress LC3B-II production, while knockdown of yes-associated protein expression reduces BCL-2 protein levels and increases LC3B-II protein levels in SW620 cells, therefore enhancing autophagy (Jin et al 2021). BCL-2 is a downstream effector for yes-associated protein to inhibit autophagy (Jin et al 2021). BCL-2 proteins have also been found to play an important role in tumor formation and multidrug resistance (Qian et al 2022).
3 groups based on primary function make up the BCL-2 family of proteins: anti-apoptotic proteins, pro-apoptotic pore-formers, and pro-apoptotic BH3-only proteins (Kale et al 2018). All BCL-2 family proteins contain a BH3 domain, which is one of four BH domains involved in interactions between these proteins (Kale et al 2018).
The anti-apoptotic proteins (BCL-2, BCL-XL, BCL-W, MCL-1, BFL-1/A1) contain all 4 BH domains and adopt a highly conserved tertiary structure farming a hydrophobic BH3 domain-binding groove that acts as a receptor for BH3 domains of other family members (Kale et al 2018).
The pro-apoptotic pore-forming proteins (BAX, BACK, BOK) also contain all 4 BH3 domains and adopt a highly conserved tertiary structure which forms a hydrophobic BH3 domain-binding groove which acts as a receptor or BH3 domains of other family members (Kale et al 2018).
The pro-apoptotic BH3-only proteins (BAD, BID, BIK, BIM, BMF, HRK, NOXA, PUMA, etc.) contain only the BH3 domain as the name suggests, are unstructured in solution (with the exception of BID), and consist of 2 groups: activator proteins, and sensitizer proteins (Kale et al 2018).
Mitochondrial outer membrane permeabilization is initiated when the BH3 domain of activator BH3-only proteins binds to the BH3 domain-binding groove in BAX/BAK, activating these proteins and eliciting a series of conformation changes that result in homo-oligomerization and pore formation within the mitochondrial outer membrane (Kale et al 2018). The BH3 domain-binding groove in anti-apoptotic proteins binds the BH3 domains of the pore-forming proteins and the activator BH3-only proteins, sequestering them and inhibiting their function (Kale et al 2018). The BH3 domain of sensitizer BH3-only proteins binds to the BH3 domain-binding groove of anti-apoptotic proteins, which inactivates them (Kale et al 2018). Most of these interactions occur at, on, or within the mitochondrial outer membrane, and the abundance and interactions between the anti-apoptotic and pro-apoptotic proteins determine mitochondrial outer membrane permeabilization, which inevitably leads to apoptosis (Kale et al 2018).
It has been found that autophagy activation is accompanied by a reduction in BCL-2 protein levels, and suppression of BCL-2 function further enhances autophagic activity (Xu et al 2013).
7. Discussion
There are many gaps in the research pertaining to these topics including gaps in glial cell and autophagy understanding. New evidence suggests that the regenerative potential of glial cells in the adult mammalian central nervous system can be expanded (Stenudd et al 2022).
Because astrocytes are so broadly and extensively involved in the central nervous system development and function, damage or deterioration to astrocytes disturbs the blood-brain barrier, synapse formation and stabilization, signaling efficiency, energy metabolism, and fluid homeostasis, causing serious damage to the central nervous system (Wei et al 2023). Astrocyte dysfunction implicates them in several neurological disorders, including Alzheimer’s Disease, epilepsy, Huntington's Disease, Parkinson's disease, multiple sclerosis, amyotrophic lateral sclerosis, and Alexander Disease (Wei et al 2023). In Alzheimer’s Disease, abnormal beta-amyloid plaques form due to increasing cleavage of amyloid precursor protein b beta and gamma secretases (Wei et al 2023). This accumulation of amyloid plaques causes astrocytes to begin deteriorating, whereas astrocytes degrade and clear amyloid plaques in a healthy central nervous system (Wei et al 2023). Too many astrocytes also interfere with adrenergic signaling, affecting homeostasis and signaling support and worsening the cognitive deficits observed in Alzheimer's Disease (Wei et al 2023). In epilepsy and Huntington’s Disease, the ability of astrocytes to regulate potassium, water, and glutamate levels in the extracellular fluid within the central nervous system is impaired (Wei et al 2023). In Parkinson’s Disease, astrocytes can initiate neuronal death and can trigger demyelinating disease (Wei et al 2023). Both Alexander Disease and inherited forms of amyotrophic lateral sclerosis stem from errors in astrocyte genetics (Wei et al 2023).
In microglia research, there is a sufficient lack of human-based experiments, and much of the information is obtained through animal experiments (Prinz et al 2019).
In oligodendrocyte research, understanding is still lacking in the area of oligodendrocyte development in adulthood, role in disease, maturation in developmental myelination (Elbaz et al 2019). In diseases, oligodendrocyte lineage cells can give rise to Schwann cells, or become antigen presenting cells (Elbaz et al 2019). Recently, posti-mitotic oligodendroctyes have been shown to participate in remyelination of the central nervous system, and adult-born myelin sheath have been found to be shorter and thinner than myelin sheath that are deposited during development (Elbaz et al 2019). The transcriptional and post-transcriptional controls of these phenomena are poorly understood (Elbaz et al 2019).
Some methods to further study the human nervous system include the generation of human microglia from embryonic stem cells, brain organoids, assembloids, 3D brain cultures, and xenotransplantation (Prinz et al 2019). Multi-omics approaches at the single-cell level, novel animal models, fate mapping and imaging tools, patient derived microglial cells, among other techniques will also aid in nervous system research (Prinz et al 2019).
In autophagy-related research, there has been growing evidence that mammalian lysosomal microautophagy plays a role in the degradation of a variety of substrates, such as endoplasmic reticulum domains containing misfolded collagen or the translocon complex, lipid droplet, or the transmembrane protein on the limiting membrane of lysosomes, but how these degradative substrates are recognized by lysosomes remains unknown (Kuchitsu et al 2023).
8. Sources
- 1. Valentini, E., D’Aguanno, S., Di Martile, M., Montesano, C., Ferraresi, V., Patsilinakos, A., Sabatino, M., Antonini, L., Chiacchiarini, M., Valente, S., Mai, A., Colotti, G., Ragno, R., Trisciuoglio, D., & Del Bufalo, D. (2022). Targeting the anti-apoptotic Bcl-2 family proteins: machine learning virtual screening and biological evaluation of new small molecules. Theranostics, 12(5), 2427–2444. https://doi.org/10.7150/thno.64233
- 2. Ramanathan, A., Parvatikar, A., Chennubhotla, S. C., Mei, Y., & Sinha, S. C. (2020). Transient Unfolding and Long-Range Interactions in Viral BCL2 M11 Enable Binding to the BECN1 BH3 Domain. Biomolecules, 10(9), 1308. https://doi.org/10.3390/biom10091308
- 3. Xu, W., Anwaier, A., Ma, C., Liu, W., Tian, X., Palihati, M., Hu, X., Qu, Y., Zhang, H., & Ye, D. (2021). Multi-omics reveals novel prognostic implication of SRC protein expression in bladder cancer and its correlation with immunotherapy response. Annals of Medicine (Helsinki)/Annals of Medicine, 53(1), 596–610. https://doi.org/10.1080/07853890.2021.1908588
- 4. Ma, A., Mu, Y., Wei, Z., Sun, M., Li, J., Jiang, H., Zhu, C., & Chen, X. (2024). SRSF10 regulates migration of neural progenitor cells and granule cells and affects the formation of dentate gyrus during the development of mouse hippocampus. Neuroscience. https://doi.org/10.1016/j.neuroscience.2024.06.012
- 5. Shahid, S. (2023, May 25). Dentate gyrus. Kenhub. Retrieved July 3, 2024, from https://www.kenhub.com/en/library/anatomy/dentate-gyrus
- 6. Nazari-Serenjeh, F., Sadeghi, M., Azizbeigi, R., Semizeh, H., Mazaheri, S., Haghparast, A., & Haghparast, A. (2024). Blocking the dopaminergic receptors within the hippocampal dentate gyrus reduced analgesic responses induced by restraint stress in the formalin test. Behavioural Brain Research, 463, 114914. https://doi.org/10.1016/j.bbr.2024.114914
- 7. Liu, X., Chen, J., Du, Y., Tian, Q., Wang, L., Li, W., Liu, G., Tan, Q., Wang, J., & Deng, X. (2024). The changes of neurogenesis in the hippocampal dentate gyrus of SAMP8 mice and the effects of acupuncture and moxibustion. Brain Research, 148814. https://doi.org/10.1016/j.brainres.2024.148814
- 8. Dieni, C. V., Gonzalez, J. C., & Overstreet-Wadiche, L. (2019). Multifaceted circuit functions of adult-born neurons. F1000Research, 8, 1998. https://doi.org/10.12688/f1000research.20642.1
- 9. Bihelek, N. (2019, January 7). Modulation of Adult Hippocampal Neurogenesis in ZnT3 Knockout Mice. https://prism.ucalgary.ca/items/e52c8482-89ce-4fed-997d-6d4f5e1d137a
- 10. Trinchero, M. F., Herrero, M., & Schinder, A. F. (2019). Rejuvenating the Brain With Chronic Exercise Through Adult Neurogenesis. Frontiers in Neuroscience, 13. https://doi.org/10.3389/fnins.2019.01000
- 11. MSEd, K. C. (2023, October 27). Neurons and Their Role in the Nervous System. Verywell Mind. https://www.verywellmind.com/what-is-a-neuron-2794890
- 12. Hirsch, L. (2022, July). Central Nervous System: The Brain and Spinal Cord (for Parents). Retrieved July 20, 2024, from https://kidshealth.org/en/parents/central-nervous-system.html#:~:text=The%20central%20nervous%20system%20is,that%20run%20throughout%20the%20body.
- 13. Wei, C., Guo, Y., Ci, Z., Li, M., Zhang, Y., & Zhou, Y. (2024). Advances of Schwann cells in peripheral nerve regeneration: From mechanism to cell therapy. Biomedicine & Pharmacotherapy, 175, 116645. https://doi.org/10.1016/j.biopha.2024.116645
- 14. Bechler, M. E., Byrne, L., & Ffrench-Constant, C. (2015). CNS Myelin Sheath Lengths Are an Intrinsic Property of Oligodendrocytes. CB/Current Biology, 25(18), 2411–2416. https://doi.org/10.1016/j.cub.2015.07.056
- 15. MSEd, K. C. (2022, November 17). All-or-None Law for Nerves and Muscles. Verywell Mind. https://www.verywellmind.com/what-is-the-all-or-none-law-2794808
- 16. Salzer, J. L. (2015). Schwann Cell Myelination. Cold Spring Harbor Perspectives in Biology, 7(8), a020529. https://doi.org/10.1101/cshperspect.a020529
- 17. Purves, D., Augustine, G. J., Fitzpatrick, D., Katz, L. C., LaMantia, A. S., McNamara, J. O., & Williams, S. M. (2001). Neuroglial Cells. Neuroscience - NCBI Bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK10869/
- 18. Allen, N. J., & Lyons, D. A. (2018). Glia as architects of central nervous system formation and function. Science, 362(6411), 181–185. https://doi.org/10.1126/science.aat0473
- 19. Allen, N. J., & Eroglu, C. (2017). Cell Biology of Astrocyte-Synapse Interactions. Neuron, 96(3), 697–708. https://doi.org/10.1016/j.neuron.2017.09.056
- 20. Rasband, M. N. (2016). Glial Contributions to Neural Function and Disease. Molecular & Cellular Proteomics, 15(2), 355–361. https://doi.org/10.1074/mcp.r115.053744
- 21. Ngoc, K. H., Jeon, Y., Ko, J., & Um, J. W. (2024). Multifarious astrocyte–neuron dialog in shaping neural circuit architecture. Trends in Cell Biology. https://doi.org/10.1016/j.tcb.2024.05.002
- 22. Wei, D. C., & Morrison, E. H. (2023, May 1). Histology, Astrocytes. StatPearls - NCBI Bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK545142/#:~:text=Astrocytes%20are%20a%20subtype%20of,barrier%2C%20and%20promoting%20synapse%20formation.
- 23. Munro, D. A., Bestard-Cuche, N., McQuaid, C., Chagnot, A., Shabestari, S. K., Chadarevian, J. P., Maheshwari, U., Szymkowiak, S., Morris, K., Mohammad, M., Corsinotti, A., Bradford, B., Mabbott, N., Lennen, R. J., Jansen, M. A., Pridans, C., McColl, B. W., Keller, A., Blurton-Jones, M., . . . Priller, J. (2024). Microglia protect against age-associated brain pathologies. Neuron. https://doi.org/10.1016/j.neuron.2024.05.018
- 24. Bozidis, C. (2024, March 19). Microglia. Kenhub. Retrieved July 22, 2024, from https://www.kenhub.com/en/library/physiology/microglia
- 25. Prinz, M., Jung, S., & Priller, J. (2019). Microglia Biology: One Century of Evolving Concepts. Cell, 179(2), 292–311. https://doi.org/10.1016/j.cell.2019.08.053
- 26. Muzio, L., Viotti, A., & Martino, G. (2021). Microglia in Neuroinflammation and Neurodegeneration: From Understanding to Therapy. Frontiers in Neuroscience, 15. https://doi.org/10.3389/fnins.2021.742065
- 27. Nayak, D., Roth, T. L., & McGavern, D. B. (2014). Microglia Development and Function. Annual Review of Immunology, 32(1), 367–402. https://doi.org/10.1146/annurev-immunol-032713-120240
- 28. Kwon, H. S., & Koh, S. H. (2020). Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Translational Neurodegeneration, 9(1). https://doi.org/10.1186/s40035-020-00221-2
- 29. Luo, T., Liang, Y., Chen, L., Sun, H., & Chen, Q. (2024). Effect of quercetin on the progression of breast cancer in mice with chronic stress by regulating the polarization of microglia. ScienceDirect. https://doi.org/10.1016/j.jff.2024.106294
- 30. Elbaz, B., & Popko, B. (2019). Molecular Control of Oligodendrocyte Development. Trends in Neurosciences, 42(4), 263–277. https://doi.org/10.1016/j.tins.2019.01.002
- 31. Barateiro, A., Brites, D., & Fernandes, A. (2016). Oligodendrocyte Development and Myelination in Neurodevelopment: Molecular Mechanisms in Health and Disease. Current Pharmaceutical Design, 22(6), 656–679. https://doi.org/10.2174/1381612822666151204000636
- 32. Zhou, B., Zhu, Z., Ransom, B. R., & Tong, X. (2020). Oligodendrocyte lineage cells and depression. Molecular Psychiatry, 26(1), 103–117. https://doi.org/10.1038/s41380-020-00930-0
- 33. Albors, A. R., Singer, G. A., Llorens-Bobadilla, E., Frisén, J., May, A. P., Ponting, C. P., & Storey, K. G. (2023). An ependymal cell census identifies heterogeneous and ongoing cell maturation in the adult mouse spinal cord that changes dynamically on injury. Developmental Cell, 58(3), 239-255.e10. https://doi.org/10.1016/j.devcel.2023.01.003
- 34. Frederico, B., Martins, I., Chapela, D., Gasparrini, F., Chakravarty, P., Ackels, T., Piot, C., Almeida, B., Carvalho, J., Ciccarelli, A., Peddie, C. J., Rogers, N., Briscoe, J., Guillemot, F., Schaefer, A. T., Saúde, L., & Sousa, C. R. E. (2022). DNGR-1-tracing marks an ependymal cell subset with damage-responsive neural stem cell potential. Developmental Cell, 57(16), 1957-1975.e9. https://doi.org/10.1016/j.devcel.2022.07.012
- 35. Delgehyr, N., Meunier, A., Faucourt, M., Grau, M. B., Strehl, L., Janke, C., & Spassky, N. (2015). Ependymal cell differentiation, from monociliated to multiciliated cells. In Methods in cell biology (pp. 19–35). https://doi.org/10.1016/bs.mcb.2015.01.004
- 36. Stenudd, M., Sabelström, H., Llorens-Bobadilla, E., Zamboni, M., Blom, H., Brismar, H., Zhang, S., Basak, O., Clevers, H., Göritz, C., Barnabé-Heider, F., & Frisén, J. (2022). Identification of a discrete subpopulation of spinal cord ependymal cells with neural stem cell properties. Cell Reports, 38(9), 110440. https://doi.org/10.1016/j.celrep.2022.110440
- 37. Song, M. H., Song, M. H., & Song, M. H. (2024). Hijacking autophagy for infection by flaviviruses. Virus Research, 347, 199422. https://doi.org/10.1016/j.virusres.2024.199422
- 38. Galhuber, M., & Thedieck, K. (2024). ODE-based Models of Signaling Networks in Autophagy. Current Opinion in Systems Biology, 100519. https://doi.org/10.1016/j.coisb.2024.100519
- 39. Mehrbod, P., Ande, S. R., Alizadeh, J., Rahimizadeh, S., Shariati, A., Malek, H., Hashemi, M., Glover, K. K. M., Sher, A. A., Coombs, K. M., & Ghavami, S. (2019). The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence, 10(1), 376–413. https://doi.org/10.1080/21505594.2019.1605803
- 40. Kuchitsu, Y., Mukai, K., Uematsu, R., Takaada, Y., Shinojima, A., Shindo, R., Shoji, T., Hamano, S., Ogawa, E., Sato, R., Miyake, K., Kato, A., Kawaguchi, Y., Nishitani-Isa, M., Izawa, K., Nishikomori, R., Yasumi, T., Suzuki, T., Dohmae, N., . . . Taguchi, T. (2023b). STING signalling is terminated through ESCRT-dependent microautophagy of vesicles originating from recycling endosomes. Nature Cell Biology, 25(3), 453–466. https://doi.org/10.1038/s41556-023-01098-9
- 41. Li, X., He, S., & Ma, B. (2020). Autophagy and autophagy-related proteins in cancer. Molecular Cancer, 19(1). https://doi.org/10.1186/s12943-020-1138-4
- 42. Tran, S., Fairlie, W. D., & Lee, E. F. (2021). BECLIN1: Protein Structure, Function and Regulation. Cells, 10(6), 1522. https://doi.org/10.3390/cells10061522
- 43. Salminen, A., Kaarniranta, K., Kauppinen, A., Ojala, J., Haapasalo, A., Soininen, H., & Hiltunen, M. (2013). Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Progress in Neurobiology, 106–107, 33–54. https://doi.org/10.1016/j.pneurobio.2013.06.002
- 44. Kang, R., Zeh, H. J., Lotze, M. T., & Tang, D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death and Differentiation, 18(4), 571–580. https://doi.org/10.1038/cdd.2010.191
- 45. Li, X., Yang, K. B., Chen, W., Mai, J., Wu, X. Q., Sun, T., Wu, R. Y., Jiao, L., Li, D. D., Ji, J., Zhang, H. L., Yu, Y., Chen, Y. H., Feng, G. K., Deng, R., Li, J. D., & Zhu, X. F. (2021). CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy, 17(12), 4323–4340. https://doi.org/10.1080/15548627.2021.1912270
- 46. Sun, Y., Yao, X., Zhang, Q. J., Zhu, M., Liu, Z. P., Ci, B., Xie, Y., Carlson, D., Rothermel, B. A., Sun, Y., Levine, B., Hill, J. A., Wolf, S. E., Minei, J. P., & Zang, Q. S. (2018). Beclin-1-Dependent Autophagy Protects the Heart During Sepsis. Circulation, 138(20), 2247–2262. https://doi.org/10.1161/circulationaha.117.032821
- 47. Hwang, H. J., Ha, H., Lee, B. S., Kim, B. H., Song, H. K., & Kim, Y. K. (2022). LC3B is an RNA-binding protein to trigger rapid mRNA degradation during autophagy. Nature Communications, 13(1). https://doi.org/10.1038/s41467-022-29139-1
- 48. Fan, S., Yue, L., Wan, W., Zhang, Y., Zhang, B., Otomo, C., Li, Q., Lin, T., Hu, J., Xu, P., Zhu, M., Tao, H., Chen, Z., Li, L., Ding, H., Yao, Z., Lu, J., Wen, Y., Zhang, N., . . . Luo, C. (2021). Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein. Angewandte Chemie, 60(50), 26105–26114. https://doi.org/10.1002/anie.202109464
- 49. Tanida, I., Ueno, T., & Kominami, E. (2004). LC3 conjugation system in mammalian autophagy. International Journal of Biochemistry & Cell Biology, 36(12), 2503–2518. https://doi.org/10.1016/j.biocel.2004.05.009
- 50. Tanida, I., Ueno, T., & Kominami, E. (2008). LC3 and Autophagy. In Methods in molecular biology (pp. 77–88). https://doi.org/10.1007/978-1-59745-157-4_4
- 51. Beccari, S., Sierra-Torre, V., Valero, J., Pereira-Iglesias, M., García-Zaballa, M., Soria, F. N., De Las Heras-Garcia, L., Carretero-Guillen, A., Capetillo-Zarate, E., Domercq, M., Huguet, P. R., Ramonet, D., Osman, A., Han, W., Dominguez, C., Faust, T. E., Touzani, O., Pampliega, O., Boya, P., . . . Sierra, A. (2023). Microglial phagocytosis dysfunction in stroke is driven by energy depletion and induction of autophagy. Autophagy, 19(7), 1952–1981. https://doi.org/10.1080/15548627.2023.2165313
- 52. Rusten, T. E., & Stenmark, H. (2010). p62, an autophagy hero or culprit? Nature Cell Biology, 12(3), 207–209. https://doi.org/10.1038/ncb0310-207
- 53. Ashraf, U., Ding, Z., Deng, S., Ye, J., Cao, S., & Chen, Z. (2021). Pathogenicity and virulence of Japanese encephalitis virus: Neuroinflammation and neuronal cell damage. Virulence, 12(1), 968–980. https://doi.org/10.1080/21505594.2021.1899674
- 54. Wakida, N. M., Lau, A. L., Nguyen, J., Cruz, G. M. S., Fote, G. M., Steffan, J. S., Thompson, L. M., & Berns, M. W. (2022). Diminished LC3-Associated Phagocytosis by Huntington’s Disease Striatal Astrocytes. Journal of Huntington’s Disease, 11(1), 25–33. https://doi.org/10.3233/jhd-210502
- 55. Zhou, T., Li, Y., Li, X., Zeng, F., Rao, Y., He, Y., Wang, Y., Liu, M., Li, D., Xu, Z., Zhou, X., Du, S., Niu, F., Peng, J., Mei, X., Ji, S. J., Shu, Y., Lu, W., Guo, F., . . . Peng, B. (2022). Microglial debris is cleared by astrocytes via C4b-facilitated phagocytosis and degraded via RUBICON-dependent noncanonical autophagy in mice. Nature Communications, 13(1). https://doi.org/10.1038/s41467-022-33932-3
- 56. Zhang, K., Wang, F., Zhai, M., He, M., Hu, Y., Feng, L., Li, Y., Yang, J., & Wu, C. (2023). Hyperactive neuronal autophagy depletes BDNF and impairs adult hippocampal neurogenesis in a corticosterone-induced mouse model of depression. Theranostics, 13(3), 1059–1075. https://doi.org/10.7150/thno.81067
- 57. Shaid, S., Brandts, C. H., Serve, H., & Dikic, I. (2012). Ubiquitination and selective autophagy. Cell Death and Differentiation, 20(1), 21–30. https://doi.org/10.1038/cdd.2012.72
- 58. Lamark, T., Svenning, S., & Johansen, T. (2017). Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays in Biochemistry, 61(6), 609–624. https://doi.org/10.1042/ebc20170035
- 59. Galluzzi, L., & Green, D. R. (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell, 177(7), 1682–1699. https://doi.org/10.1016/j.cell.2019.05.026
- 60. Filali-Mouncef, Y., Hunter, C., Roccio, F., Zagkou, S., Dupont, N., Primard, C., Proikas-Cezanne, T., & Reggiori, F. (2021). The ménage à trois of autophagy, lipid droplets and liver disease. Autophagy, 18(1), 50–72. https://doi.org/10.1080/15548627.2021.1895658
- 61. Szebényi, K., Wenger, L. M. D., Sun, Y., Dunn, A. W. E., Limegrover, C. A., Gibbons, G. M., Conci, E., Paulsen, O., Mierau, S. B., Balmus, G., & Lakatos, A. (2021). Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nature Neuroscience, 24(11), 1542–1554. https://doi.org/10.1038/s41593-021-00923-4
- 62. Liu, J., Liu, W., & Yang, H. (2018). Balancing Apoptosis and Autophagy for Parkinson’s Disease Therapy: Targeting BCL-2. ACS Chemical Neuroscience, 10(2), 792–802. https://doi.org/10.1021/acschemneuro.8b00356
- 63. Dong, Y., Chen, H., Gao, J., Liu, Y., Li, J., & Wang, J. (2019). Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. Journal of Molecular and Cellular Cardiology/Journal of Molecular and Cellular Cardiology, 136, 27–41. https://doi.org/10.1016/j.yjmcc.2019.09.001
- 64. Kale, J., Osterlund, E. J., & Andrews, D. W. (2017). BCL-2 family proteins: changing partners in the dance towards death. Cell Death and Differentiation, 25(1), 65–80. https://doi.org/10.1038/cdd.2017.186
- 65. Pattingre, S., & Levine, B. (2006). Bcl-2 Inhibition of Autophagy: A New Route to Cancer? Cancer Research, 66(6), 2885–2888. https://doi.org/10.1158/0008-5472.can-05-4412
- 66. Jin, L., Chen, Y., Cheng, D., He, Z., Shi, X., Du, B., Xi, X., Gao, Y., & Guo, Y. (2021). YAP inhibits autophagy and promotes progression of colorectal cancer via upregulating Bcl-2 expression. Cell Death and Disease, 12(5). https://doi.org/10.1038/s41419-021-03722-8
- 67. Qian, S., Wei, Z., Yang, W., Huang, J., Yang, Y., & Wang, J. (2022). The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Frontiers in Oncology, 12. https://doi.org/10.3389/fonc.2022.985363
- 68. Xu, H. D., Wu, D., Gu, J. H., Ge, J. B., Wu, J. C., Han, R., Liang, Z. Q., & Qin, Z. H. (2013). The Pro-Survival Role of Autophagy Depends on Bcl-2 Under Nutrition Stress Conditions. PloS One, 8(5), e63232. https://doi.org/10.1371/journal.pone.0063232
Document information
Published on 30/07/24
Submitted on 22/07/24
Licence: CC BY-NC-SA license
Share this document
Keywords
claim authorship
Are you one of the authors of this document?