Abstract
Background
Factors causing resistance to renal denervation (RDN) for treatment of arterial hypertension are not known. In the current study, we sought to determine mechanisms involved in responsiveness to renal denervation therapy in patients with difficult-to-control and resistant hypertension.
Methods and results
We evaluated the differential CpG methylation of genes in blood samples isolated from patients of a recently described cohort of responders or non-responders to renal denervation using microarray technique and measured protein levels of identified downstream effectors in blood samples of these patients by ELISA.
Our analysis revealed up to 6103 methylation sites differing significantly between non-responders and responders to renal denervation therapy. Software based analysis showed several of these loci to be relevant for arterial hypertension and sympathetic nervous activity. Particularly, genes involved in glutamate synthesis, degradation and glutamate signaling pathways were differently methylated between both groups. For instance, genes for glutamate dehydrogenase 1 and 2 central to glutamate metabolism, genes for ionotropic (AMPA, NMDA) and metabotropic glutamate receptors as well as glutamate transporters revealed significant differences in methylation correlating with responsiveness to RDN. To underline their potential relevance for responsiveness to RDN, we measured plasma protein levels of norepinephrine, a downstream effector of the glutamate receptor pathway, which were significantly lower in non-responders to RDN.
Conclusions
The present study describes novel molecular targets potentially contributing to reduction of blood pressure after RDN in some patients. Identifying patients with a high responsiveness to RDN could contribute to an individualized therapy in drug resistant hypertension.
Abbreviations
ABPM, ambulatory blood pressure monitoring;AF, atrial fibrillation;BP, blood pressure;BRS, baroreflex sensitivity;GFR, glomerular filtration rate;NE, norepinephrine;RDN, renal denervation therapy;PVI, pulmonal vein isolation
Keywords
Epigenetics;Drug resistant hypertension;Renal denervation therapy
1. Introduction
Catheter-based renal sympathetic denervation (RDN) is a novel treatment option in patients with resistant hypertension [1]. However, RDN is an invasive procedure with the inherent risks of side effects. The net clinical benefit of this procedure is subject of ongoing clinical trials [2], and clear determinants to predict beneficial outcome of this intervention are warranted.
During RDN, sympathetic outflow to the kidney is reduced by inflicting damage to the renal peri-vascular nervous system [3]. Since the first description of interventional renal denervation, several experimental and clinical studies suggested a beneficial effect on arterial hypertension [4]; [5] ; [6]. RDN was also suggested to have effects on pathophysiological settings other than arterial hypertension such as ventricular arrhythmias, glucose metabolism or insulin sensitivity [7] ; [8]. While the underlying mechanisms are not entirely characterized, in the setting of catecholamine dependent ventricular tachycardia, modulation of cardio-cardiac reflexes of sympathetic origin may account for the observed effects [9]; [10]; [11] ; [12]. Now, experimental and clinical research tries to define indicators, which can predict RDN success, and clinical markers to identify patients responding to the procedure.
Although arterial hypertension is a multifactorial disease, dysregulation of sympathetic/parasympathetic activity is a central mechanism of its pathophysiology. Accordingly, sympathetic nerve activity is considered a major contributor to the pathophysiology of arterial hypertension [13]. Dysbalance of sympathetic activity may be a result of changes in signaling tailored by epigenetic changes in expression of molecules associated with the sympathetic signaling transduction [14]. For example in patients suffering from postural tachycardia syndrome, it was demonstrated that expression of the norepinephrine transporter is inhibited by suppressive histone-methylation [15]. These pathophysiological considerations render the sympathetic regulatory system a potential candidate to understand mechanisms of resistance to blood pressure regulation. Recently, we described a novel approach to identify patients as responders or non-responders to RDN by measuring baroreflex sensitivity (BRS) obtained from non-invasive recordings as surrogate measure for underlying sympathetic tone [16]. We were able to demonstrate that impaired BRS was a strong and significant predictor of response to RDN [16]. Epigenetic regulation is discussed as one of the emerging mechanisms affecting arterial hypertension [17]. To uncover possible targets, which affect responsiveness to RDN, we performed microarray analysis of responders and non-responders in our BRS patient collective and determined differential methylation possibly resulting in decreased DNA transcription.
2. Materials and methods
2.1. Ethics
Patients' samples were obtained after patients gave written and informed consent. The study was approved by the local ethics committee and conforms with the ethical principles outlined in the Declaration of Helsinki.
2.2. Patients
The study includes 5 randomly selected responders as well as age-, gender-, glomerular filtration rate (GFR)-, treatment- and blood pressure-matched non-responders of a previously described cohort with resistant arterial hypertension undergoing RDN [16]. Briefly, inclusion criteria were defined as follows: patients were 18 years or older, had an office based BP of ≥ 160 mm Hg, a mean systolic BP of ≥ 130 mm Hg on ambulatory blood pressure monitoring (ABPM) were on at least 3 antihypertensive drugs, had a GFR of ≥ 45 ml/min− 1/1.73 m− 2 and no known secondary cause of high BP except sleep apnea or chronic kidney disease. Identically to our previous report [16], response to RDN was defined as reduction of mean systolic arterial blood pressure of 10 mm Hg or greater 6 months after RDN.
2.3. Sample collection
In patients undergoing RDN, blood was taken from the arterial femoral access at the beginning of the procedure. Blood samples were then immediately transferred to blood collection tubes containing EDTA (Sarstedt, Nuembrecht, Germany). Blood samples for DNA analysis were immediately frozen as a whole and stored at − 80 °C. For isolation of EDTA plasma, samples were spun down at room temperature at 1250 × g for 10 min. Plasma samples were then collected and stored at − 80 °C.
2.4. Norepinephrine (NE) enzyme linked immunosorbent assays (ELISA)
EDTA plasma samples were thawed on ice, resuspended and analyzed using a NE ELISA (Labor Diagnostika Nord, Nordhorn, Germany) following the manufacturers protocol. All samples were analyzed in duplicate and measured on a microplate reader (Bio-Rad Laboratories, Munich, Germany) at 450 nm. n = 5 samples were analyzed for responders and nonresponders.
2.5. BRS-PRSA
Assessment of cardiac BRS was carried out as described before [16]. Briefly, patients underwent simultaneous 30-min high-resolution electrocardiographic recordings (1.6 kHz in orthogonal XYZ leads) and noninvasive continuous arterial BP monitoring using a finger hotoplethysmographic device (Finapres; TNO-TPD Biomedical Instrumentation, Amsterdam, the Netherlands). The recordings were analyzed according to standardized conditions by an experienced technician blinded to the clinical status of the patient. Cardiac BRS was assessed from the series of RR intervals and systolic BP values by phase-rectified signal averaging (PRSA). For BRS-PRSA, increases of systolic BP (BP) are identified within the BP time series. Subsequently, segments of RR intervals around BP are identified and averaged. The resulting bivariate PRSA signal shows RR oscillations related to increases of systolic BP, whereas heart rate variability due to other causes is eliminated by the averaging process.
2.6. Methylation array
Total DNA was isolated from frozen 2.8 ml EDTA blood samples (n = 5) for responders and nonresponders respectively using the QIAamp DNA Mini Kit (QIAgen, Hilden, Germany) according to the manufacturers protocol.
500 ng of total DNA were bisulfite converted for methylation analysis according to the manufacturers protocol using the EZ DNA Methylation Kit (Zymo Research, Irvine CA, USA). Bisulfite converted DNA was then applied to Infinium Human Methylation450 BeadChip (Illumina, San Diego CA, USA) and analyzed using an Illumina iScan microarray scanner (Illumina, San Diego CA, USA) according to the manufacturers protocol. Genes differing in methylation with high significance were further analyzed for association with regulatory pathways, networks and disease. Data analysis was performed using GenomeStudio software (Illumina, San Diego CA, USA) and QIAGENs Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The pathways shown were generated through the use of QIAGENs Ingenuity Pathway Analysis (IPA®,QIAGEN Redwood City, www.qiagen.com/ingenuity). Methylation analysis was performed at the Department of Medical Genetics, Microarray Facility, Tuebingen, Germany.
2.7. Statistical analysis
Preliminary statistical analysis of methylation data was carried out in the programming environment “R” using the packages “lumi” and “methylumi”. Resulting β-values were normalized and converted to M-values using logit-transformation. M-values were analyzed in “R” using the “limma” package. For methylation array statistical analysis, genes were considered significantly different between both groups (responder vs. nonresponders), if β-values of at least an absolute 10% were observed. Methylation sites meeting this requirement and showing an uncorrected p-value of p ≤ 0.05 were considered for pathway analysis. For ELISA and BRS-PRSA values, statistical analysis was performed with SPSS (version 22, IBM Deutschland GmbH, Ehningen, Germany). Concentrations obtained in the ELISA-assay were compared using Mann–Whitney U-test.
3. Results
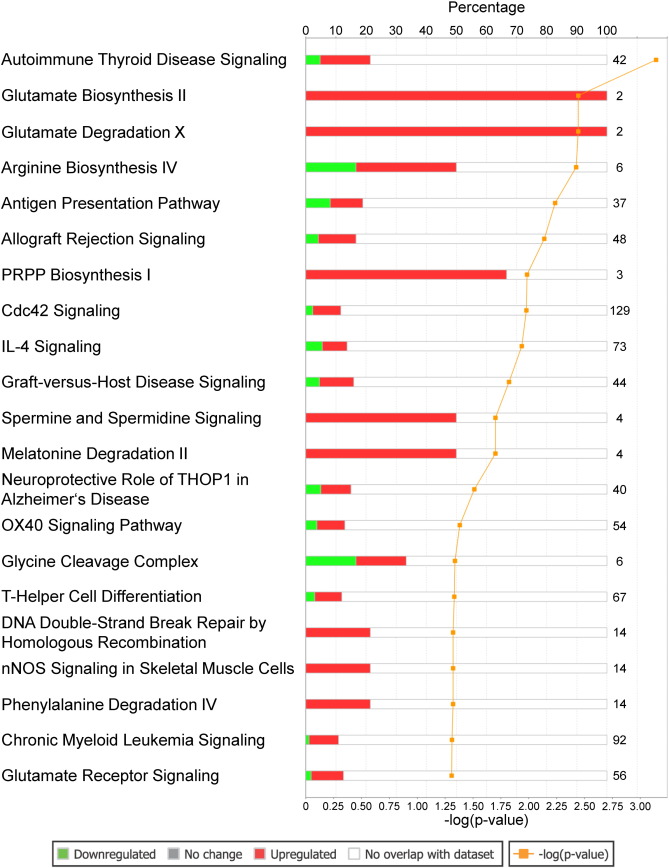
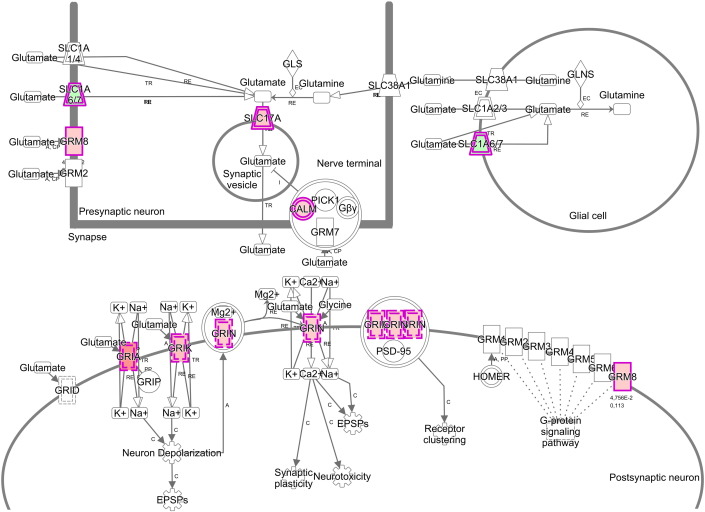
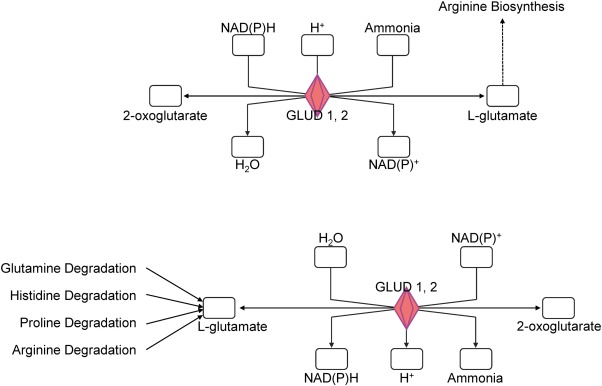
The patients' characteristics of “responders or nonresponders to RDN” are depicted in Table 1. There was no significant difference regarding factors important for the pathogenesis of arterial hypertension such as gender, body mass index or diabetes. Comparing patients responding to RDN with nonresponders, we were able to identify a large number of sites with differential CpG-methylation. Criteria for selection of methylation sites for further analysis of genes are given in supplemental Fig. 1. Genes differentially methylated between the 2 groups with a minimum difference in methylation of at least 10% absolute and confidence interval p ≤ 0.05 were considered for software based pathway analysis. Analysis of 6103 sites meeting these predefined criteria of significance revealed a group of 56 differentially methylated genes to be potentially associated with hypertension (Table 2). Further analysis indicated a potential modification of pathways linked to hypertension such as glutamate biosynthesis and degradation (both p < 0.005) as well as glutamate signaling (p < 0.05) (Fig. 1). Interestingly, also parts of non-expected pathways such as genes involved in antigen presentation or IL-4 signaling were differentially methylated (Fig. 1). In glutamate signaling, the affected genes encode both ionotropic and metabotropic glutamate receptors, located mainly in the postsynaptic neuron (Fig. 2). Furthermore, we also identified glutamate transporters and a calcium dependent kinase, mostly located in the presynaptic neuron being differentially methylated between responders and non-responders to RDN (Fig. 2, Table 3). Genes involved in glutamate biosynthesis and degradation can be classified as glutamate dehydrogenases mediating both processes (Fig. 3, Table 3).
| Variable | Responder (n = 5) | Non-responder (n = 5) | p-Value |
|---|---|---|---|
| Age | 57 +/− 14 | 55 +/− 21 | 0.684 |
| Female gender | 3/5 (60%) | 1/5 (20%) | 0.524 |
| Body mass index | 33 +/− 5 | 29 +/− 4 | 0.721 |
| Diabetes mellitus | 2/5 (40%) | 3/5 (60%) | 1.000 |
| # antihypertensive med. baseline | 4.2 +/− 1.1 | 5.0 +/− 0.7 | 0.067 |
| # antihypertensive med. 6 months | 4.8 +/− 1.1 | 4.2 +/− 1.1 | 0.623 |
| Systolic BP baseline (mm Hg) | 161 +/− 11 | 149 +/− 17 | 0.664 |
| Systolic BP 6 months (mm Hg) | 135 +/− 6 | 145 +/− 12 | 0.198 |
Patient characteristics.
| Gene symbol | Encoded protein/RNA | β-Diff |
|---|---|---|
| ACE2 | Angiotensin-converting enzyme 2 | − 0.114 |
| ADRA1A | Alpha-1A adrenergic receptor | − 0.199 |
| ADRA2C | Alpha-2C adrenergic receptor | − 0.120 |
| AFF3 | AF4/FMR2 family member 3 | 0.516 |
| ALOX12 | Arachidonate 12-lipoxygenase, 12S-type | − 0.369 |
| ARHGEF10 | Rho guanine nucleotide exchange factor 10 | − 0.103 |
| ATP2B3 | Plasma membrane calcium-transporting ATPase 3 | − 0.177 |
| CA5B | Carbonic anhydrase 5B, mitochondrial | 0.138 |
| CACNA1F | Voltage-dependent L-type calcium channel subunit alpha-1F | 0.143 |
| CBX7 | Chromobox protein homolog 7 | − 0.193 |
| CCDC81 | Coiled-coil domain-containing protein 81 | 0.112 |
| CELF4 | CUGBP Elav-like family member 4 | 0.226 |
| CHM | Rab proteins geranylgeranyltransferase component A1 | 0.126 |
| CYB5B | Cytochrome b5 type B | − 0.120 |
| DNAJB6 | DnaJ homolog subfamily B member 6 | − 0.543 |
| FANCB | Fanconi anemia group B protein | 0.165 |
| FHL1 | Four and a half LIM domain protein 1 | 0.136 |
| GALNT18 | Polypeptide N-acetylgalactosaminyltransferase 18 | 0.119 |
| GPIHBP1 | Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 | − 0.118 |
| GYPC | Glycophorin-C | 0.135 |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 | 0.134 |
| JARID2 | Protein Jumonji | − 0.114 |
| LINC00087 | Long intergenic non-protein coding RNA 87 | 0.102 |
| LMO3 | LIM domain only protein 3 | 0.119 |
| LONRF3 | LON peptidase N-terminal domain and RING finger protein 3 | 0.132 |
| MAOA | Amine oxidase [flavin-containing] A | 0.143 |
| MXRA5 | Matrix-remodeling-associated protein 5 | − 0.160 |
| NEGR1 | Neuronal growth regulator 1 | − 0.318 |
| NKRF | NF-kappa-B-repressing factor | 0.148 |
| NMNAT3 | Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 3 | − 0.309 |
| NOTCH4 | Neurogenic locus notch homolog protein 4 | 0.440 |
| PDE11A | Dual 3,5-cyclic-AMP and -GMP phosphodiesterase 11A | − 0.259 |
| PDZD4 | PDZ domain-containing protein 4 | 0.107 |
| PIGA | Phosphatidylinositol N-acetylglucosaminyltransferase subunit A | 0.110 |
| PNMA3 | Paraneoplastic antigen Ma3 | 0.283 |
| PRRG1 | Transmembrane gamma-carboxyglutamic acid protein 1 | 0.106 |
| PTGFR | Prostaglandin F2-alpha receptor | 0.184 |
| RAB1A | Ras-related protein Rab-1A | 0.225 |
| RBFOX1 | RNA binding protein fox-1 homolog 1 | 0.105 |
| RBFOX3 | RNA binding protein fox-1 homolog 3 | − 0.175 |
| SCN4B | Sodium channel subunit beta-4 | 0.248 |
| SLC39A8 | Zinc transporter ZIP8 | 0.203 |
| SLC6A1 | Sodium- and chloride-dependent GABA transporter 1 | 0.118 |
| SLC6A2 | Sodium-dependent noradrenaline transporter | − 0.147 |
| SLC7A3 | Cationic amino acid transporter 3 | 0.226 |
| SLITRK4 | SLIT and NTRK-like protein 4 | 0.142 |
| SRPX | Sushi repeat-containing protein SRPX | 0.108 |
| THBS2 | Thrombospondin-2 | − 0.115 |
| TIMP1 | Metalloproteinase inhibitor 1 | 0.135 |
| TMEM47 | Transmembrane protein 47 | 0.254 |
| TMSB15B | Thymosin beta-15B | 0.144 |
| WDR27 | WD repeat-containing protein 27 | 0.114 |
| XPNPEP2 | Xaa-Pro aminopeptidase 2 | − 0.132 |
| ZDHHC15 | Palmitoyltransferase ZDHHC15 | 0.133 |
| ZNF614 | Zinc finger protein 614 | 0.234 |
Genes showing differential methylation in responders vs nonresponders (p ≤ 0.05) and associated with hypertension according to software based analysis (p ≤ 0.05); ß-Diff = methylation (responder) − methylation (nonresponder).
|
|
|
Fig. 1. Association of differentially methylated genes with signaling pathways in responders vs nonresponders to RDN. Numbers at the end of bars indicate number of genes in the underlying pathway, filled bars indicate percentage of differentially methylated genes; green color indicates lower methylation, red color stronger methylation in the responder group. Statistical significance (p < 0.05) and, thus, non-random association of genes to pathways is indicated by the yellow curve showing − log (p-value). |
|
|
|
Fig. 2. Molecules involved in glutamate receptor signaling and their synaptic location. Icons in red color indicate stronger methylation for samples isolated from responders to RDN, genes in green indicate lower methylation. CALM: calmodulin. GRIA = glutamate receptor, ionotropic AMPA. GRIK = glutamate receptor, ionotropic kainate. GRIN = glutamate receptor, ionotropic NMDA. GRM8 = glutamate receptor, metabotropic 8. SLC1A 6/7 = solute carrier family 1, member 6/7. SLC17 = solute carrier family 17. |
| Glutamate signaling | ||
|---|---|---|
| Gene symbol | Classification | β-Diff |
| CAMK4 | Kinases | 0.110 |
| GRIA3 | AMPA receptor | 0.148 |
| GRIN2B | NMDA receptor | 0.113 |
| GRIK4 | GRIK4 | 0.145 |
| GRM8 | Class 3 metabotrope receptor | 0.113 |
| SLC1A7 | Transporter | − 0.140 |
| SLC17A7 | Transporter | 0.125 |
| Glutamate biosynthesis and degradation | ||
| GLUD1 | Glutamate dehydrogenase | 0.183 |
| GLUD2 | Glutamate dehydrogenase | 0.117 |
List of genes involved in glutamate signaling (p ≤ 0.05) or glutamate biosynthesis and degradation (p ≤ 0.005) differentially methylated in responders vs nonresponders to RDN and their classification; ß-Diff = methylation (responder) − methylation (nonresponder).
|
|
|
Fig. 3. Pathway analysis defined by software based meta analysis. The graphic depicts molecules involved in glutamate biosynthesis (top) and degradation (bottom); red color indicates stronger methylation for corresponding genes in responder samples. GLUD 1, 2 = glutamate dehydrogenase 1, 2. |
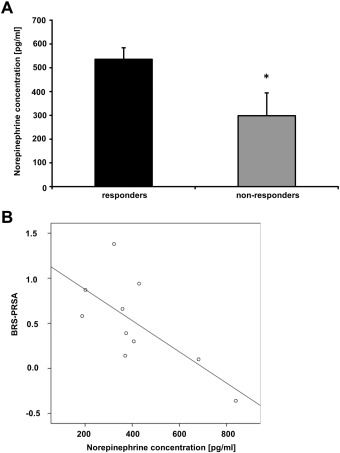
As these differential methylations may cause differences in sympathetic function changes in NE plasma levels [18] ; [19], we measured NE levels in responders and nonresponders. Interestingly, we detected a significantly lower concentration of norepinephrine (Fig. 4A) in the blood of patients not responding to RDN compared to responders (p < 0.05). Moreover, plasma NE levels showed an inverse correlation with BRS-PRSA signals (Pearson coefficient r = − 0.70, p ≤ 0.05, Fig. 4B).
|
|
|
Fig. 4. Differential CpG methylation is associated with reduced norepinephrine levels in non-responders to RDN. (A) ELISA analysis of EDTA plasma samples. Levels of blood norepinephrine (NE) were measured in plasma samples by ELISA, n = 5 for responders and n = 5 for non-responders to RDN, p < 0.05. Error bars show standard error of mean. (B) Significant inverse correlation between levels of plasma NE and BRS-PRSA. Pearson coefficient r = − 0.70, p < 0.05. |
4. Discussion
Patients suffering from arterial hypertension, which are beyond any further promising medical treatment eventually will develop complications such as chronic cardiovascular disease, stroke or acute myocardial infarction associated with high morbidity and mortality. Therefore, novel alternative treatment options such as interventional renal denervation are urgently needed to improve quality of life and clinical outcome in these patients. In a recently described patient cohort, we assessed responsiveness to renal denervation by cardiac BRS to select optimal candidates for RDN and to identify patients, in which RDN should be avoided [16]. This is of particular clinical relevance, as a recent randomized trial showed no substantial benefit for RDN over the sham procedure [20].
Here, we characterized underlying mechanisms of “response or resistance” to renal denervation using microarray chip technology. For instance, we analyzed patients classified as “non-responders” or “responders” to RDN based on their decrease in ABPM with regard to regulated pathways potentially affecting the sympathetic nervous system. We i) identified pathways and a number of genes involved in hypertension differentially methylated between responders and non-responders to RDN, ii) observed that genes important for neuronal glutamate signal transduction or glutamate biosynthesis/degradation were differentially methylated in responders vs nonresponders and iii) measured significantly lower levels of norepinephrine, a central downstream effector of the glutamate receptor pathway, in blood samples of non-responders to RDN.
The sympathetic nervous system controls blood pressure through glucosensitive, thermosensitive and barosensitive relays responsible for the control of short and long-term blood pressure. These sensors control norepinephrine release from organs such as the heart, kidneys and from a subset of adrenal chromaffine cells, and as a consequence vessel resistance by constriction of arterioles [21]. Moreover, they play a role in renin secretion, sodium reabsorption in renal tubules and renal blood flow contributing to the long-term control of BP [22]. These pathophysiological considerations and the strong abundance of efferent sympathetic nerves located around the kidney arteries, the approach of RDN to treat patients with arterial hypertension resistant to regular medical antihypertensive treatment was proposed. In clinical trials, it was demonstrated that RDN can attenuate efferent renal sympathetic activity, which was demonstrated by assessment of NE spillover (Symplicity HTN1) [6] or dampening of central sympathetic activity as verified by effects on muscle sympathetic nerve activity and cardiac BRS [6] ; [23]. As we detected differing plasma NE levels, it is tempting to speculate that this is one of the reasons, why responders are more susceptible to modulation of sympathetic drive by renal denervation therapy than non-responders. In other words, in patients with lower NE levels, the “target” for renal denervation therapy may be missing.
Some of the differentially methylated genes associated with hypertension shown in Table 2 could indicate possible physiological differences between responders and non-responders resulting in different RDN outcome. The gene ACE2 encoding the angiotensin I converting enzyme 2, a homolog of ACE, showed increased methylation in the non-responders group. ACE2 functions as a monocarboxypeptidase with the primary function of degrading Ang II to Ang 1–7, which in turn acts through its recepetor Mas changing the balance of the renin angiotensin system from vasoconstriction to vasodilation [24]. As a consequence, lower expression of ACE2 indicated by a higher degree of methylation would cause the control of BP to be prone to higher BP. Interestingly, a reduced expression of ACE2 was found in 3 different models of hypertension in rats compared to normotensive controls [25]. Additionally, the genes ADRA1A encoding for the adrenoreceptor Alpha 1 A, and ADRA2C encoding for the adrenoreceptor Alpha 2C could indicate a difference in sympathetic signaling between responders and non-responders. Both genes showed a higher methylation in the nonresponder group. In sympathetic signaling, the alpha 1 receptors are located on the postsynaptic neuron and bind released NE to mediate effects on the target tissue, whereas alpha 2 receptors are located on the presynaptic neuron providing negative feedback after binding NE and inhibiting further NE release [26].
Based on the consideration that other diseases such as cardiac arrhythmias feature strong sympathetic activity and malfunction of its self-regulation, researchers and clinicians started to extend application of RDN to these entities [7]; [8]; [9]; [27]; [28] ; [29]. In a more complex clinical setting, combination of RDN with pulmonal vein isolation (PVI) in patients suffering from drug resistant hypertension and refractory atrial fibrillation (AF) showed the expected reduction in BP as well as significantly better results in reducing AF than in a control group only undergoing PVI [29]. The authors discuss the possibility that this is due to a combination of reduced mechanical stress caused by lower blood pressure and decrease of central sympathetic output as a result of reducing afferent renal nervous sympathetic input [29].
In conclusion, this study provides potential novel mechanisms underlying non-responsiveness to RDN and, thus, offers potential molecular targets to further characterize and approach difficult-to-control and resistant hypertension.
5. Study limitations
Our study has limitations such as the low number of patients analyzed by microarray chip technology. Thus, we cannot exclude a possible selection bias. All patients were treated with the interventional device used in the SIMPLICITY trials, and we cannot exclude that a significant reduction of blood pressure would have been achieved in the “non-responder” group using an alternative device. Furthermore, insufficient attachment of the catheter to the renal artery wall may have caused non-responsiveness to RDN. Also mechanisms other than sympathetic dependent ones such as factors of the immune system [30] may be involved in the lack of efficacy of RDN and will have to be evaluated in future studies.
The following are the supplementary data related to this article.
|
|
|
Supplemental fig. 1. Criteria for selection of methylation sites for further analysis of genes. Genes were categorized according to significance thresholds. Genes corresponding at least to the third category (p < 0.05) were considered for this publication. 6103 differentially methylated sites resulted in 1335 genes, which were used for further analysis. 479475 methylation sites were below the statistical threshold. Differences in number of methylation sites and number of genes analyzed are a result of multiple methylation sites belonging to the same gene. ß-Diff = beta-difference. |
Author contributions
F.E. carried out the sample collection, ELISA measurements, analyzed the data and contributed to writing the manuscript. C.S.Z. conceptualized the study and revised the manuscript. K.D.R. collected patient data and critically revised the manuscript. G.J. performed statistical analysis on DNA methylation data and provided support on interpretation of the results. S.G.M, C.E., M.G., and A.B. performed data analysis and critically reviewed the manuscript. J.P. analyzed correlations for BRS-PRSA with NE levels. H.F.L. conceived and designed the study, analyzed data and wrote the manuscript.
Grant support
The study was supported by grants from the Volkswagen Foundation (Lichtenberg program) and the German Heart Foundation to HFL.
Conflict of interest
None.
Acknowledgments
We acknowledge the excellent technical assistance of Sarah Gekeler and the help with microarray analysis by Dr. Michael Bonin.
References
- [1] M. Bohm, D. Linz, C. Ukena, M. Esler, F. Mahfoud; Renal denervation for the treatment of cardiovascular high risk-hypertension or beyond?; Circ. Res., 115 (2014), pp. 400–409
- [2] A. Persu, S. Kjeldsen, J.A. Staessen, M. Azizi; Renal denervation for treatment of hypertension: a second start and new challenges; Curr. Hypertens. Rep., 18 (2016), p. 6
- [3] M.P. Schlaich, P.A. Sobotka, H. Krum, R. Whitbourn, A. Walton, M.D. Esler; Renal denervation as a therapeutic approach for hypertension: novel implications for an old concept; Hypertension, 54 (2009), pp. 1195–1201
- [4] R.H. Smithwick, J.E. Thompson; Splanchnicectomy for essential hypertension; results in 1,266 cases; J. Am. Med. Assoc., 152 (1953), pp. 1501–1504
- [5] D.M. Morrissey, V.S. Brookes, W.T. Cooke; Sympathectomy in the treatment of hypertension; review of 122 cases; Lancet, 1 (1953), pp. 403–408
- [6] H. Krum, M. Schlaich, R. Whitbourn, P.A. Sobotka, J. Sadowski, K. Bartus, et al.; Catheter-based renal sympathetic denervation for resistant hypertension: a multicentre safety and proof-of-principle cohort study; Lancet, 373 (2009), pp. 1275–1281
- [7] C. Ukena, A. Bauer, F. Mahfoud, J. Schreieck, H.R. Neuberger, C. Eick, et al.; Renal sympathetic denervation for treatment of electrical storm: first-in-man experience; Clin. Res. Cardiol., 101 (2012), pp. 63–67
- [8] F. Mahfoud, M. Schlaich, I. Kindermann, C. Ukena, B. Cremers, M.C. Brandt, et al.; Effect of renal sympathetic denervation on glucose metabolism in patients with resistant hypertension: a pilot study; Circulation, 123 (2011), pp. 1940–1946
- [9] P.J. Schwartz; Cardiac sympathetic denervation to prevent life-threatening arrhythmias; Nat. Rev. Cardiol., 11 (2014), pp. 346–353
- [10] P.J. Schwartz, H.L. Stone; Left stellectomy in the prevention of ventricular fibrillation caused by acute myocardial ischemia in conscious dogs with anterior myocardial infarction; Circulation, 62 (1980), pp. 1256–1265
- [11] A.A. Wilde, Z.A. Bhuiyan, L. Crotti, M. Facchini, G.M. De Ferrari, T. Paul, et al.; Left cardiac sympathetic denervation for catecholaminergic polymorphic ventricular tachycardia; N. Engl. J. Med., 358 (2008), pp. 2024–2029
- [12] B. Thorens; Brain glucose sensing and neural regulation of insulin and glucagon secretion; Diabetes Obes. Metab., 13 (Suppl. 1) (2011), pp. 82–88
- [13] G.F. DiBona; The sympathetic nervous system and hypertension: recent developments; Hypertension, 43 (2004), pp. 147–150
- [14] M. Esler, N. Eikelis, M. Schlaich, G. Lambert, M. Alvarenga, D. Kaye, et al.; Human sympathetic nerve biology: parallel influences of stress and epigenetics in essential hypertension and panic disorder; Ann. N. Y. Acad. Sci., 1148 (2008), pp. 338–348
- [15] R. Bayles, K.N. Harikrishnan, E. Lambert, E.K. Baker, A. Agrotis, L. Guo, et al.; Epigenetic modification of the norepinephrine transporter gene in postural tachycardia syndrome; Arterioscler. Thromb. Vasc. Biol., 32 (2012), pp. 1910–1916
- [16] C.S. Zuern, C. Eick, K.D. Rizas, S. Bauer, H. Langer, M. Gawaz, et al.; Impaired cardiac baroreflex sensitivity predicts response to renal sympathetic denervation in patients with resistant hypertension; J. Am. Coll. Cardiol., 62 (2013), pp. 2124–2130
- [17] M. Liang, A.W. Cowley Jr., D.L. Mattson, T.A. Kotchen, Y. Liu; Epigenomics of hypertension; Semin. Nephrol., 33 (2013), pp. 392–399
- [18] H. Kannan, Y. Hayashida, H. Yamashita; Increase in sympathetic outflow by paraventricular nucleus stimulation in awake rats; Am. J. Physiol. Regul. Integr. Comp. Physiol., 256 (1989), p. R1325-R30
- [19] T. Katafuchi, Y. Oomura, M. Kurosawa; Effects of chemical stimulation of paraventricular nucleus on adrenal and renal nerve activity in rats; Neurosci. Lett., 86 (1988), pp. 195–200
- [20] D.L. Bhatt, D.E. Kandzari, W.W. O'Neill, R. D'Agostino, J.M. Flack, B.T. Katzen, et al.; A controlled trial of renal denervation for resistant hypertension; N. Engl. J. Med., 370 (2014), pp. 1393–1401
- [21] W. Janig, H.J. Habler; Neurophysiological analysis of target-related sympathetic pathways — from animal to human: similarities and differences; Acta Physiol. Scand., 177 (2003), pp. 255–274
- [22] G.F. DiBona, U.C. Kopp; Neural control of renal function; Physiol. Rev., 77 (1997), pp. 75–197
- [23] M.P. Schlaich, P.A. Sobotka, H. Krum, E. Lambert, M.D. Esler; Renal sympathetic-nerve ablation for uncontrolled hypertension; N. Engl. J. Med., 361 (2009), pp. 932–934
- [24] G.I. Rice, D.A. Thomas, P.J. Grant, A.J. Turner, N.M. Hooper; Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism; Biochem. J., 383 (2004), pp. 45–51
- [25] M.A. Crackower, R. Sarao, G.Y. Oudit, C. Yagil, I. Kozieradzki, S.E. Scanga, et al.; Angiotensin-converting enzyme 2 is an essential regulator of heart function; Nature, 417 (2002), pp. 822–828
- [26] S.Z. Langer; Presynaptic regulation of the release of catecholamines; Pharmacol. Rev., 32 (1980), pp. 337–362
- [27] A. Kannan, R.I. Medina, N. Nagajothi, S. Balamuthusamy; Renal sympathetic nervous system and the effects of denervation on renal arteries; World J. Cardiol., 6 (2014), pp. 814–823
- [28] D. Linz, A. van Hunnik, C. Ukena, S. Ewen, F. Mahfoud, S.H. Schirmer, et al.; Renal denervation: effects on atrial electrophysiology and arrhythmias; Clin. Res. Cardiol., 103 (2014), pp. 765–774
- [29] E. Pokushalov, A. Romanov, G. Corbucci, S. Artyomenko, V. Baranova, A. Turov, et al.; A randomized comparison of pulmonary vein isolation with versus without concomitant renal artery denervation in patients with refractory symptomatic atrial fibrillation and resistant hypertension; J. Am. Coll. Cardiol., 60 (2012), pp. 1163–1170
- [30] M.J. Ryan; An update on immune system activation in the pathogenesis of hypertension; Hypertension, 62 (2013), pp. 226–230
Document information
Published on 19/05/17
Submitted on 19/05/17
Licence: Other
Share this document
claim authorship
Are you one of the authors of this document?