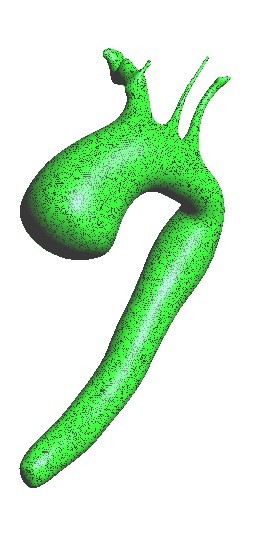m (Cinmemj moved page Draft Samper 526155048 to CIMNE 2009b) |
|||
| (27 intermediate revisions by the same user not shown) | |||
| Line 106: | Line 106: | ||
<big>* 1D-Vessel model of whole human body. General information coupled to specific 2D or 3D studies. </big> | <big>* 1D-Vessel model of whole human body. General information coupled to specific 2D or 3D studies. </big> | ||
| − | [[Image:draft_Samper_526155048-image27.png|center| | + | {| |
| − | + | |- | |
| − | [[Image:draft_Samper_526155048-image28-c.png|center| | + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image27.png|center|150px]] |
| − | + | |[[Image:draft_Samper_526155048-image28-c.png|center|150px]] | |
| − | [[Image:draft_Samper_526155048-image29.jpeg|center| | + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image29.jpeg|center|150px]] |
| − | + | |} | |
<big>* Reconstruction of real geometries starting by DICOM images.</big> | <big>* Reconstruction of real geometries starting by DICOM images.</big> | ||
| Line 121: | Line 121: | ||
<big> | <big> | ||
| − | [[Image:draft_Samper_526155048-image31.png|center| | + | [[Image:draft_Samper_526155048-image31.png|center|500px]] |
</big> | </big> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Biomaterials== | ==Biomaterials== | ||
| − | Development of biocompatible geometries for internal or external devices (stents, internal prosthesis, etc). | + | Development of biocompatible geometries for internal or external devices (stents, internal prosthesis, etc). |
<big>* Finite element for stress testes with biomaterials and medical devices.</big> | <big>* Finite element for stress testes with biomaterials and medical devices.</big> | ||
| Line 148: | Line 133: | ||
<big>* | <big>* | ||
| − | [[Image:draft_Samper_526155048- | + | [[Image:draft_Samper_526155048-image35.jpeg|center|300px]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</big> | </big> | ||
| + | |||
| + | |||
| + | <big>* New constitutive models for biomaterial and shape memory materials.</big> | ||
<big>* Parameter identifications in constitutive models of biomaterials.</big> | <big>* Parameter identifications in constitutive models of biomaterials.</big> | ||
| − | + | [[Image:draft_Samper_526155048-image37.jpeg|center|300px]] | |
| − | [[Image:draft_Samper_526155048- | + | |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {| style="text-align: center;" | ||
| + | |- | ||
| + | |[[Image:draft_Samper_526155048-image38.jpeg|center|300px]] | ||
| + | |[[Image:draft_Samper_526155048-image39.jpeg|center|300px]] | ||
| + | |- | ||
| + | |[[Image:draft_Samper_526155048-image40.jpeg|center|350px]] | ||
| + | |[[Image:draft_Samper_526155048-image36.jpeg|center|200px]] | ||
| + | |} | ||
==Artifial Intelligence== | ==Artifial Intelligence== | ||
| Line 184: | Line 170: | ||
<big>* Development of intelligent finite element methods via Al Technology</big> | <big>* Development of intelligent finite element methods via Al Technology</big> | ||
| − | + | {| style="text-align: center;" | |
| − | + | |- | |
| − | [[Image:draft_Samper_526155048- | + | |[[Image:draft_Samper_526155048-image45.jpeg|center|300px]] |
| − | + | | [[Image:draft_Samper_526155048-image44.jpeg|center|300px]] | |
| − | + | |- | |
| − | + | | [http://www.cimne.com/flood/ http://www.cimne.com/flood/] | |
| − | + | | | |
| − | [[Image:draft_Samper_526155048- | + | |} |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Neursciences== | ==Neursciences== | ||
| Line 215: | Line 197: | ||
| − | + | {| style="text-align: center;" | |
| − | + | |- | |
| − | [[Image:draft_Samper_526155048-image50.png|center|300px]] | + | |[[Image:draft_Samper_526155048-image50.png|center|300px]] |
| − | + | | style="padding-left:20px;"| [[Image:draft_Samper_526155048-image49.png|center|300px]] | |
| − | + | |} | |
| − | + | ||
| − | [[Image:draft_Samper_526155048- | + | |
| − | + | ||
==Medical-GiD== | ==Medical-GiD== | ||
| − | |||
| − | |||
| − | = | + | {| style="text-align: center; " |
| − | [[Image:draft_Samper_526155048-image53.jpeg|center|300px]] | + | |- |
| + | | [[Image:draft_Samper_526155048-image53.jpeg|center|300px]] | ||
| + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image54.png|center|300px]] | ||
| + | |[[Image:draft_Samper_526155048-image52.jpeg|center|300px]] | ||
| + | |- | ||
| + | |Magnetic Resonance (2D) | ||
| + | | 2D Detail | ||
| + | |Edition/Generation | ||
| + | |} | ||
| − | |||
| − | [[Image:draft_Samper_526155048- | + | {| style="text-align: center; " |
| + | |- | ||
| + | |[[Image:draft_Samper_526155048-image55.png|center|300px]] | ||
| + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image56.png|center|250px]] | ||
| + | |- | ||
| + | |Deformable isosurface model | ||
| + | |Meshing of heart and aorta | ||
| + | |- | ||
| + | |[[Image:draft_Samper_526155048-image58-c.jpeg|center|300px]] | ||
| + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image57-c.jpeg|center|200px]] | ||
| + | |- | ||
| + | |Meshing of heart | ||
| + | |3D heart | ||
| + | |} | ||
| − | |||
| − | |||
| − | + | <big>* Segmentation and 3D reconstruction of medical images. </big> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Image:draft_Samper_526155048-image60.png|center|300px]] | [[Image:draft_Samper_526155048-image60.png|center|300px]] | ||
| − | |||
| − | |||
<big>* Meshing of segmented geometries: creation of surface meshes or volume meshes.</big> | <big>* Meshing of segmented geometries: creation of surface meshes or volume meshes.</big> | ||
| Line 265: | Line 245: | ||
<big>* Visualization of 4D images (3D + time), creation of flux vectors and study of time developing in the image.</big> | <big>* Visualization of 4D images (3D + time), creation of flux vectors and study of time developing in the image.</big> | ||
| − | |||
| − | |||
| − | [[ | + | [[File:Draft_Samper_526155048_2501_FigIT573.png|center]] |
| − | |||
| − | |||
<big>* Anatomical real cases.</big> | <big>* Anatomical real cases.</big> | ||
| Line 279: | Line 255: | ||
<big>* Friendly platform and portability of the informatics solutions adopted.</big> | <big>* Friendly platform and portability of the informatics solutions adopted.</big> | ||
| − | |||
| − | |||
| − | |||
| − | + | {| style="text-align: center; " | |
| − | [[Image:draft_Samper_526155048-image65-c.jpeg|center|300px]] | + | |- |
| − | + | |[[Image:draft_Samper_526155048-image65-c.jpeg|center|300px]] | |
| + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image64-c.jpeg|center|300px]] | ||
| + | |[[Image:draft_Samper_526155048-image66-c.jpeg|center|300px]] | ||
| + | |} | ||
| − | + | ==Urology== | |
| − | + | ||
| − | + | ||
| − | + | <big>* Finite Element Method for the simulation of the urinary bladder and its parts like the destrusor (little smooth muscle)</big> | |
| − | <big>* | + | |
| − | Finite Element Method for the simulation of the urinary bladder and its parts like the destrusor (little smooth muscle)</big> | + | |
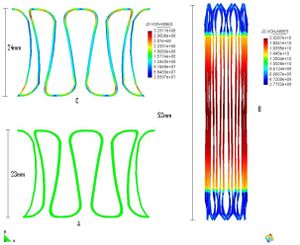
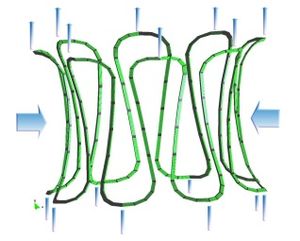
<big>* Study of biological materials and its multi-scale hierarchy, creation of simplificated models with classical nonlinear continuum mechanics theory. </big> | <big>* Study of biological materials and its multi-scale hierarchy, creation of simplificated models with classical nonlinear continuum mechanics theory. </big> | ||
| − | |||
[[Image:draft_Samper_526155048-image69.jpeg|center|300px]] | [[Image:draft_Samper_526155048-image69.jpeg|center|300px]] | ||
| − | <big>* | + | |
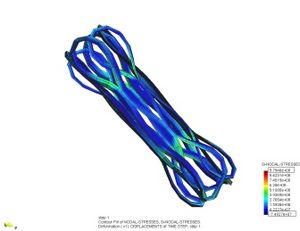
| + | <big>* Characterization of destrusor-tissue model is based in the representation (based on hyperelastic matrix, and viscoelastic fibres)</big> | ||
| + | |||
| + | |||
[[Image:draft_Samper_526155048-image70.jpeg|center|300px]] | [[Image:draft_Samper_526155048-image70.jpeg|center|300px]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <big>* Analisys of the interaction between bladder wall with urine modelled via the Particle Finite Element Method (PFEM) </big> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | [[Image:draft_Samper_526155048-image73.png|center|300px]] | ||
| + | |style="padding:15px;"|[[Image:draft_Samper_526155048-image68.jpeg|center|300px]] | ||
| + | |} | ||
==Pre and Post Proccesing== | ==Pre and Post Proccesing== | ||
| Line 317: | Line 294: | ||
<big>* Development of methods for generating structure and unstructured meshes.</big> | <big>* Development of methods for generating structure and unstructured meshes.</big> | ||
| − | <big>* | + | <big>* Development of input data technology for large scale computational problems.</big> |
| − | + | ||
| + | <big>* Graphical visualization techniques for large scale simulation problems.</big> | ||
| + | |||
| + | <big>* Generation of input data for finite element analysis from medical images.</big> | ||
| + | |||
| + | <big>* Meshless methods for parameterization of geometries for shape optimization problems.</big> | ||
[[Image:draft_Samper_526155048-image75.png|center|300px]] | [[Image:draft_Samper_526155048-image75.png|center|300px]] | ||
[[Image:draft_Samper_526155048-image76.png|center|300px]] | [[Image:draft_Samper_526155048-image76.png|center|300px]] | ||
| − | |||
| − | + | [[Image:draft_Samper_526155048-image74.png|center|300px]] | |
| − | + | [[File:Draft_Samper_526155048_1299_Fig2IT573.png|center|300px]] | |
| − | |||
| − | < | + | <div class="center"> |
| − | [[Image:draft_Samper_526155048-image77.png|center| | + | [[Image:draft_Samper_526155048-image77.png|center|250px]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [http://www.gidhome.com www.gidhome.com] </div> | |
Latest revision as of 13:10, 13 June 2018
Research Lines & RTD Project in Biomedical Engineering
Computational Fluid Dynamics
Solid and Structural Biomechanics
Health Decision Support Systems
Cardiovascular System
Biomaterials
Artificial Intelligence
Neurosciences
Medical-GiD
Urology
Pre and post processing
Computational Fluid Dynamics
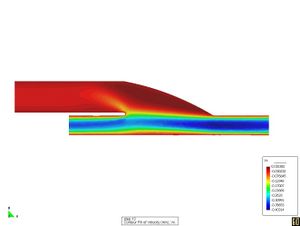
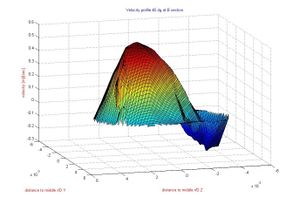
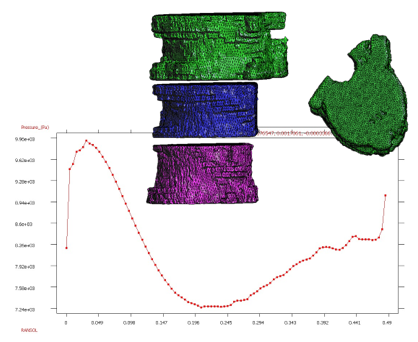
* Stabilized finite element and finite difference methods in incompressible biofluid mechanics.
* Bio-Absorption theory application in vessel structures for atheroma plack and biochemical studies.
* Finite element methods for fluid flow and analysis.
* Numerical methods applied in multidisciplinary problems in fluid biomechanics (fluid structure interaction, thermal flows, absorption theory etc).
* Coupling 3D with 2D or 1D models to improve study details.
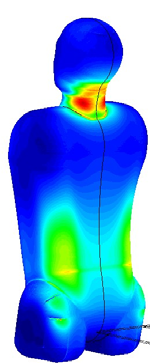
Solid and Structural Biomechanics
* Finite element methods for linear and non linear analysis of solids structures.
* Coupled problems in solid biomechanics (fluid structure interaction, thermal flows, absorption theory etc).
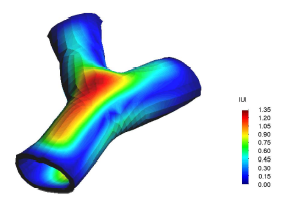
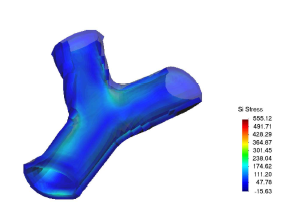
* Finite element methods for biomechanical devices analysis and prototype design (stent, prosthesis, etc).
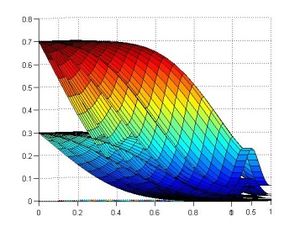
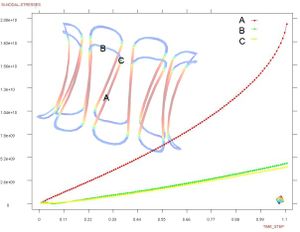
* Finite element methods analysis of solid biology structures (hearth mechanics, vessel stresses response, etc).
Health Decision Support System
* Development of intelligent platform to help physician work, informatization of routinely medical work.
* Finite element use to improve medical diagnosis and to perfect analysis processes.
* Biostatistical models applied ad hoc for several medical problems and cases.
* Bioinformatic technology solutions to coupled finite elements methods with biostatistical tools and artificial intelligence.
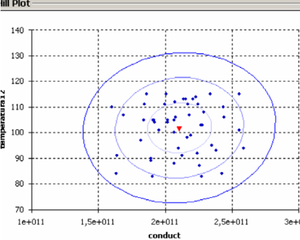
* Monte-Carlo methods for stochastic analysis in computational biomechanics and in biofluid dynamics.
* Parameter identification via stochastic methods.
* Coupling of TIC solutions, stochastic methods and finite element methods to improve and get faster medical analysis and decision
Computational Fluid Dynamics
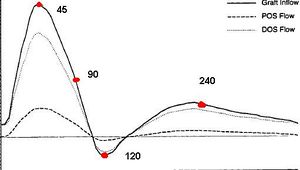
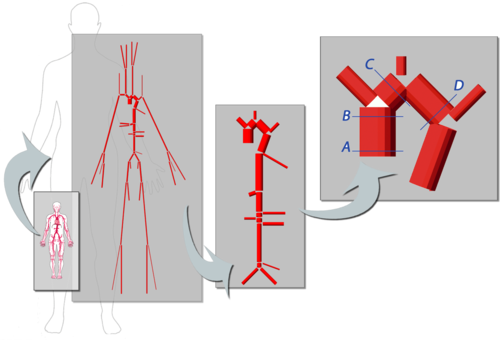
* Development of simulation platform for cardiovascular problems.
* Finite element for the simulation of problematic scenarios (aneurism, lumen obstruction, deformation, etc).
* Finite element for the study of cholesterol and platelets vessel absorption.
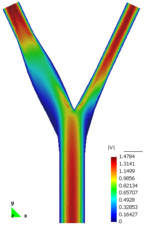
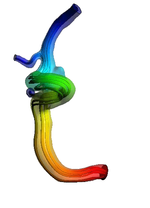
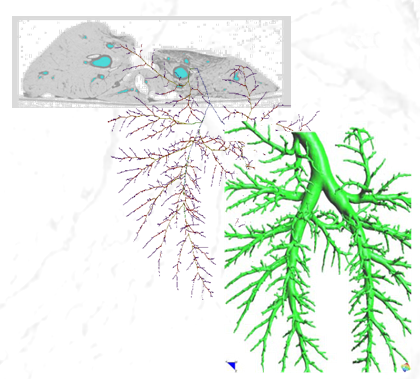
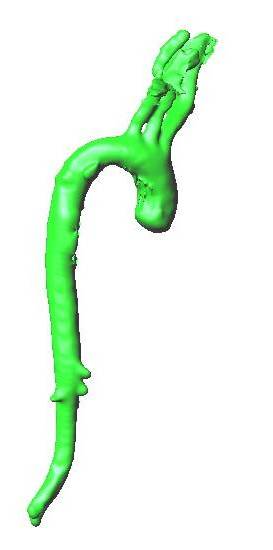
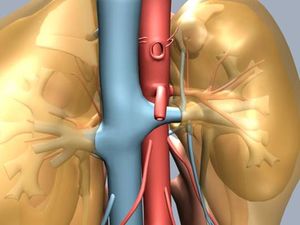
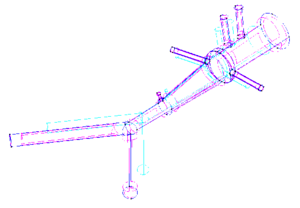
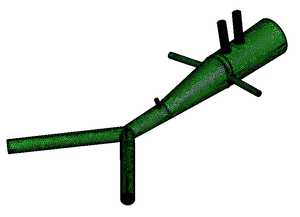
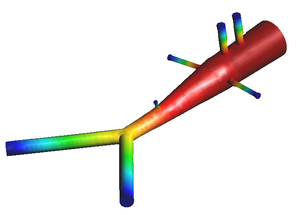
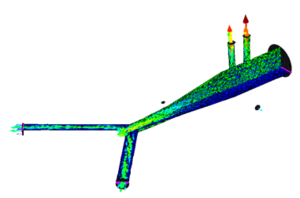
* 1D-Vessel model of whole human body. General information coupled to specific 2D or 3D studies.
* Reconstruction of real geometries starting by DICOM images.
* Automatic 2D and 3D geometries for vessel obstruction or aneurisms formation analysis.
Biomaterials
Development of biocompatible geometries for internal or external devices (stents, internal prosthesis, etc).
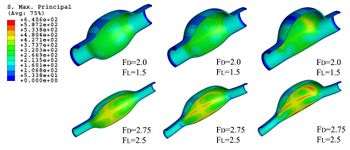
* Finite element for stress testes with biomaterials and medical devices.
* Design and study of biocompatible devices for human medical use or experimental use.
*
* New constitutive models for biomaterial and shape memory materials.
* Parameter identifications in constitutive models of biomaterials.
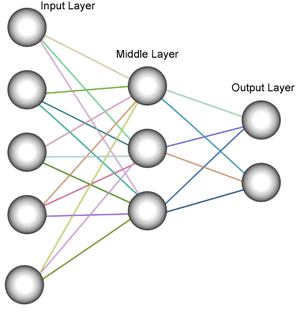
Artifial Intelligence
* Development of artificial neural networks (ANN) for optimization, inverse analysis and medical decision support fast decision taking.
* Integration of artificial neural networks (ANN) in decision support systems combining wireless sensors, computer simulations methods and artificial intelligence technology.
*
Development of artificial intelligence techniques based in agent simulations.
* Applications of artificial neural networks (ANN) technology for parameter identification in constitutive laws
* Development of intelligent finite element methods via Al Technology
| http://www.cimne.com/flood/ |
Neursciences
* Finite element methods for the analysis of brain cellular activity in pathological and physiological scenarios.
* 1D Finite element methods to study the propagations of neuronal signals in complex networks.
*
Statistical methods to fast response in biochemical brain analysis.
* Dementia diseases studies: finite element methods and bioinformatic solutions to reinforce the investigation about the causes of several brain dysfunction.
* Amyloids, Polymers and Cerebral Membrane Characterization
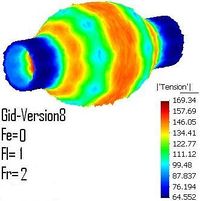
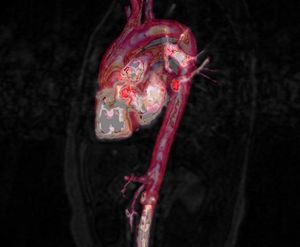
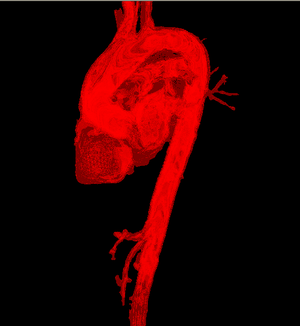
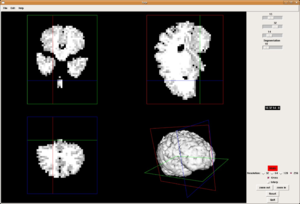
Medical-GiD
| Magnetic Resonance (2D) | 2D Detail | Edition/Generation |
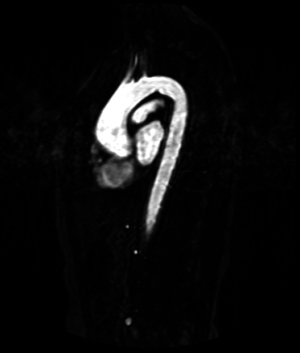
| Deformable isosurface model | Meshing of heart and aorta |
| Meshing of heart | 3D heart |
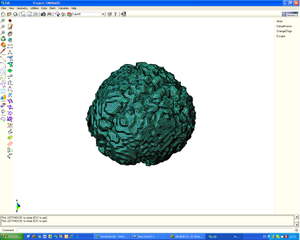
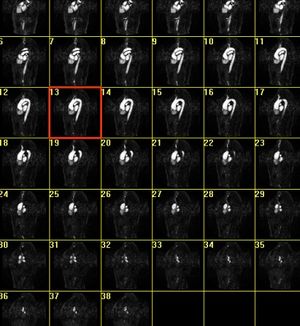
* Segmentation and 3D reconstruction of medical images.
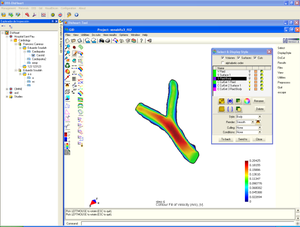
* Meshing of segmented geometries: creation of surface meshes or volume meshes.
* Visualization of 4D images (3D + time), creation of flux vectors and study of time developing in the image.
* Anatomical real cases.
* Coupling with simulation programs and with finite element methods solver.
* Friendly platform and portability of the informatics solutions adopted.
Urology
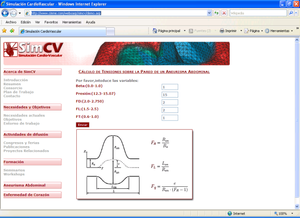
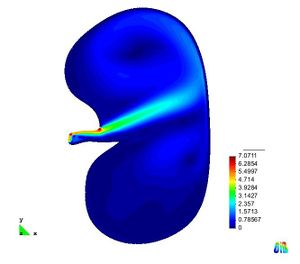
* Finite Element Method for the simulation of the urinary bladder and its parts like the destrusor (little smooth muscle)
* Study of biological materials and its multi-scale hierarchy, creation of simplificated models with classical nonlinear continuum mechanics theory.
* Characterization of destrusor-tissue model is based in the representation (based on hyperelastic matrix, and viscoelastic fibres)
* Analisys of the interaction between bladder wall with urine modelled via the Particle Finite Element Method (PFEM)
Pre and Post Proccesing
* Development and maintenance of GiD pre and post processing system (www.gidhome.com).
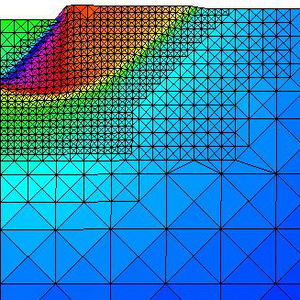
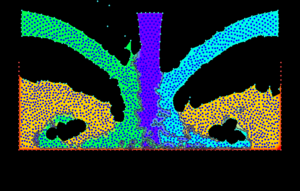
* Development of methods for generating structure and unstructured meshes.
* Development of input data technology for large scale computational problems.
* Graphical visualization techniques for large scale simulation problems.
* Generation of input data for finite element analysis from medical images.
* Meshless methods for parameterization of geometries for shape optimization problems.
Document information
Published on 01/01/2009
Licence: CC BY-NC-SA license
Share this document
Keywords
claim authorship
Are you one of the authors of this document?