(Created page with " ==Abstract== Toxoplasmosis is a worldwide zoonosis caused by the parasitic protozoan ''Toxoplasma gondii''. It can infect all warm-blooded vertebrate species and causes abo...") |
m (Scipediacontent moved page Draft Content 120485166 to Rouatbi et al 2016a) |
(No difference)
| |
Latest revision as of 10:37, 9 June 2017
Abstract
Toxoplasmosis is a worldwide zoonosis caused by the parasitic protozoan Toxoplasma gondii. It can infect all warm-blooded vertebrate species and causes abortions and birth defects in pregnant women and pregnant ewes. The objective of this study was to estimate the prevalence of infection with T. gondii in sheep meat in the region of Sidi Bouzid (central Tunisia) and Beja (northern Tunisia), the realization of a descriptive study of risk factors and the phylogenetic analyses of T. gondii. Neck muscle samples were obtained from 174 ewes and ewe lamb slaughtered in Sidi Bouzid and 150 lambs slaughtered in Beja. DNA was extracted from the samples using the Wizard® genomic DNA purification kit. A nested PCR using two pairs of primers (NN 1 and NN2, Tg-NP1 and Tg-NP2) were used to detect infection with T. gondii, which was then confirmed by sequencing. Eight T. gondii amplicons were sequenced (accession number KT896498) and deposited in GenBank. The T. gondii amplicons showed 97–100% identities with GenBank sequences. A phylogenetic tree was then constructed. The nested PCR detected T. gondii DNA in 31% of animals tested in Sidi Bouzid and 32% of lambs tested in Beja. No significant difference in the prevalence of T. gondii infection was established between the two tested regions. In both regions, no significant variation of the infection depending on age, breed and locality was found.
Introduction
The total population of sheep in Tunisia is 6.5 million (Ben Salem et al. 2011). They are among the most economically important livestock species in Tunisia and play an important role in the livelihood of resource-poor farmers. Small ruminants face important health problems with high prevalence of several diseases such as brucellosis, foot and mouth disease, border disease and several parasitic infections like fasciolosis (Akkari et al. 2011), lungworms (Lahmar et al. 2012) and gastrointestinal helminths (Akkari et al. 2012). Toxoplasma gondii infection is also common in sheep in Tunisia. This parasite is distributed worldwide infecting all warm-blooded vertebrate species, including humans. In humans, infection during pregnancy can cause congenital toxoplasmosis (Dunn et al. 1999). Horizontal transmission occurs frequently after ingestion of cysts in undercooked meat or by consumption of food contaminated with sporulated oocysts (Dubey et al. 2014). Recently, infection through goat and sheep raw milk has been confirmed (de Santana Rocha et al. 2015; Amairia et al. 2016). It has been shown that insemination, using experimentally contaminated fresh semen with different T. gondii tachyzoites concentrations was able to transmit infection to sheep, indicating the possibility of a sexual transmission route (Bezerra et al. 2014).
Toxoplasma gondii infection occurs asymptomatically in most immunocompetent hosts (Długońska 2014). Most infected humans do not show clinical signs, but transmission of T. gondii to the foetus can cause serious neurological abnormalities and ultimately death (Flatt & Shetty 2013). In some cases, the infection remains unapparent until the second or third decade of life (Jones et al. 2001). A study conducted by World Health Organization ranked foodborne toxoplasmosis as the most important foodborne parasitic disease with 10.3 million cases between 2010 and 2015 (Torgerson et al. 2015). In 2001, the overall prevalence of toxoplasmosis in girls living in the North of Tunisia was 58.4%. It rose from 24.5% at 10 years age to 52.1% at 20 years (Bouratbine et al. 2001). Prevalence in pregnant women in other African countries varies from 85.4% in Ethiopia (Gelaye et al. 2015) to 23.7% in the Governorate of El Fayoum, Egypt (Ghoneim et al. 2010).
In sheep, primary infection during pregnancy can lead to foetal resorption during early pregnancy or abortion, causing high economic losses (Johnston 1988; Buxton et al. 1991). Infection between 50 and 120 days of gestation leads to abortion, mummified foetuses or the birth of stillborn and weak lambs. After 120 days of gestation, the infection generally leads to apparently normal lambs that can survive the first week, grow normally and are immune to re-infection (Buxton & Finlayson 1986). Sheep become infected by either consuming oocysts contaminated food or vertically. The presence of cats is also a risk factor for animals’ infection.
Prevalence of T. gondii infection in sheep has been estimated around the world. In North Africa, the lowest molecular prevalence was reported in Tunisia (1.8% in the Governorate of Siliana) (Gharbi et al. 2013) and the highest one in Egypt (98.4% in the Governorate of El Fayoum) (Ghoneim et al. 2010). In Ghana, 33.2% of the studied sheep were infected (Van der Puije et al. 2000), whereas in South Africa, the overall national seroprevalence in sheep was found to be 5.6% by IFAT and 4.3% by ELISA in five studied provinces (Samra et al. 2007). In Europe, the seroprevalence values varied between 27.8% and 97% (Opsteegh et al. 2010; Leblebicier & Yildiz 2014). In Asia, prevalence varied between 17.8% in Kuwait (Alazemi 2014) and 69.9% in Bangladesh (Rahman et al. 2014). Toxoplasmosis has also been studied in several Latin American countries such as Colombia (58%), Brazil (53.3%) and Argentina (17.3%) (Perry et al. 1978; Hecker et al. 2013; Cosendey-KezenLeite et al. 2014).
The diagnosis of toxoplasmosis can be made using serological assays, isolation of the parasite in mice or amplification of parasite DNA by different molecular techniques, namely PCR, qPCR and loop-mediated isothermal amplification (LAMP). Although the LAMP technique is a sensitive, easy and least time-consuming method (Wang et al. 2013), PCR was used in this trial because the price of LAMP reagents is high.
In Tunisia, despite the importance of toxoplasmosis, few studies have been devoted to this parasite in sheep. The aim of this work was to estimate the molecular prevalence of T. gondii in two different regions of Tunisia (North and Central) to analyse the risk factors and to characterize this parasite genetically.
Material and methods
Study area
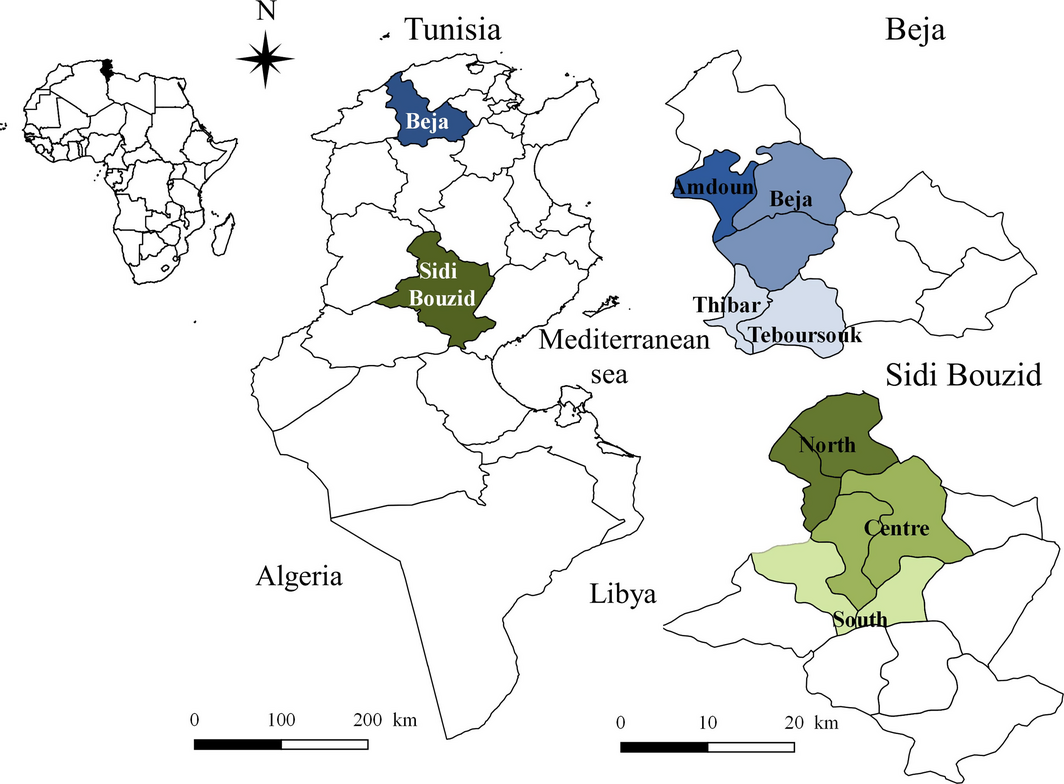
This survey was carried out in two slaughterhouses located in two Tunisian governorates Sidi Bouzid and Beja (Fig. 1). This choice of the geographical areas was based on the previous study by Gharbi et al. (2013) targeting regions with the highest and the lowest seroprevalence rates, namely the Centre and the North of the country, respectively. The district of Sidi Bouzid has a latitude of 35°02′ N, a longitude of 9°29 ′E and a mean altitude of 332 m (Dateandtime.info). Sidi Bouzid is an arid area located in Central Tunisia with a mean annual rainfall of 223 mm. The mean minimal temperature is 8.8°C in January and the mean maximum temperature is 27.6°C in July (Climate data.org). Sidi Bouzid is known as an important sheep production area with 635 000 head in 2013–2014 (Ministry of Agriculture 2014) under semi-intensive conditions relying on grazing in degraded rangeland and the main breeds are the hardy Barbarine and Queue Fine de l'Ouest. The district of Beja has a latitude of 36°43′ N, a longitude of 9°10′ E and a mean altitude of 248 m (Dateandtime.info). It is a temperate area located in North Tunisia with a mean annual rainfall of 662 mm. The mean minimal temperature is 9.3°C in January and the mean maximum temperature is 27.3°C in August (Climate data.org). The district is also known for semi-intensive sheep production under integrated crop–forage systems with a population of 369 580 heads mainly represented by the dairy Sicilo Sarde, the northern type of the Barbarine breed and several crosses between the autochthonous breeds (Ministry of Agriculture 2014). In both districts, the locality where the animal originated was noted.
|
|
|
Figure 1. Study area of Toxoplasma gondii in sheep: Northern (Beja) and Central (Sidi Bouzid) Tunisia (North Africa). |
Sample collection and preparation
During the 2013 summer season, 174 ewes and ewe lambs from the slaughterhouse of Sidi Bouzid (103 ewes and 71 ewe lambs) and 150 ewe lambs from the slaughterhouse of Beja were sampled for Toxoplasma gondii infection. Animals in the slaughterhouses came from different regions of Sidi Bouzid and Beja, and were chosen randomly. Both ewes and ewe lambs were available at this time because it was the culling period. Sheep from Sidi Bouzid were divided into four age groups: less than 1 year, between 1 and 3 years, between 4 and 5 years and finally between 6 and 10 years. Sheep from Beja were divided into four age groups: 5, 6, 7 and between 8 and 12 months. A piece of meat weighing approximately 10 g was cut from the neck of each animal using a disposable scalpel blade, placed in sterile bags and brought in a cooling box to the laboratory of parasitology of the National School of Veterinary Medicine for further processing, storage and analyses. Samples were stored at −20°C at maximum for 4 months before being used.
DNA extraction and polymerase chain reactions
DNA was extracted from 0.5 g of each meat sample using Wizard® genomic DNA purification kit (Promega, Madison, WI) according to the manufacturers instructions and then stored at −20°C until used. A PCR, targeting the 18S rRNA, was carried out in a mixture consisting of 1× PCR buffer, 2 mmol/L MgCl2, 10 μmol/L of each primer (1A and 564R), 0.2 mmol/L of each dNTP, 2U Taq polymerase (Vivantis, Chino, CA), 1.5 μL of DNA template and distilled water to a total volume of 25 μL (Wang et al. 2014) (Table 1). The DNA was amplified using the following program: 5 min denaturation at 94°C, followed by 25 cycles (94°C for 50 s and 58°C for 50 s each) and a final extension at 72°C for 10 min (Fig. 2). Three microlitres of each positive sample were amplified by a nested PCR detecting T. gondii DNA (Hurtado et al. 2001). Two PCR reactions were performed in two tubes with two programs using successively a set of two primers: external primers (NN1 and NN2) hybridizing with a T. gondii and Neospora caninum region of the ITS1 gene and internal primers (Tg-NP1 and Tg-NP2) amplifying a region of 227 bp of ITS1 gene of T. gondii (Hurtado et al. 2001) (Table 1) (Fig. 3). The two amplification rounds were carried out in 25 μL of mixture consisting of 1× PCR buffer, 200 μmol/L of each dNTP, 2 mmol/L of MgCl2, 0.5 U of Taq and 0.1 μmol/L of each external and internal primer, respectively. The DNA was amplified using the following program: 3 min denaturation at 94°C, followed by 30 cycles (94°C for 30 s, 67°C for 45 s and 72°C for 1 min for the first round and 94°C for 30 s, 53°C for 30 s and 72°C for 30 s for the second round) and a final extension at 72°C for 5 min. PCR products were examined by electrophoresis on 1.5% agarose gel stained with ethidium bromide and visualized under ultraviolet light.
| Primer specificity | Target gene | Name | Type | Primers 5′—3′ | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| Universal PCR | 18S rRNA | 1A | Forward primer | AACCTGGTTGATCCTGCCAGT | – | Wang et al. (2014) |
| 564R | Reverse primer | GGCACCAGACTTGCCCTC | ||||
| T. gondii | ITS1 rDNA | MN1 | External forward primer | CCTTTGAATCCCAAGCAAAACATGAG | Hurtado et al. (2001) | |
| MN2 | External reverse primer | GCGAGCCAAGACATCCATTGCTGA | ||||
| Tg-NP1 | Internal forward primer | GTGATAGTATCGAAAGGTAT | 227 | |||
| Tg-NP2 | Internal reverse primer | ACTCTCTCTCAAATGTTCCT |
|
|
|
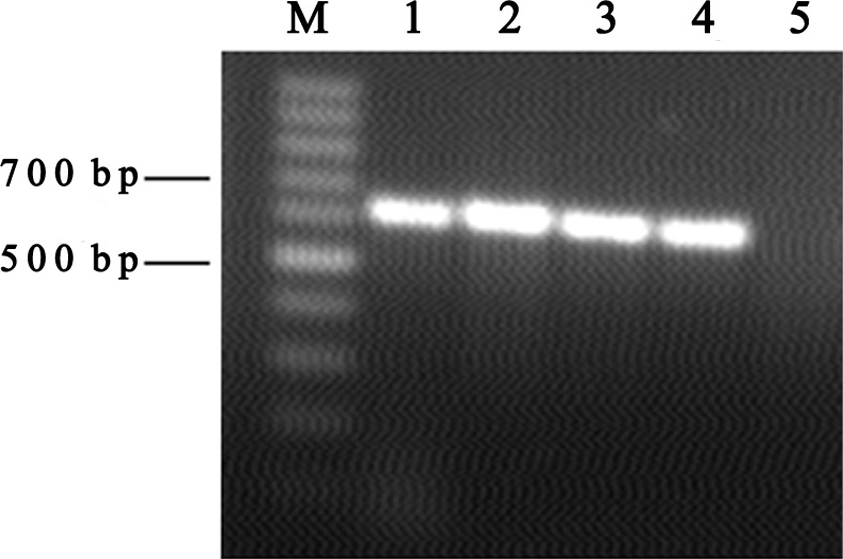
Figure 2. Agarose gel electrophoresis of universal DNA. M: 100 bp ladder; 1: positive control; 5: negative control; 2, 3, 4: positive PCR products. |
|
|
|
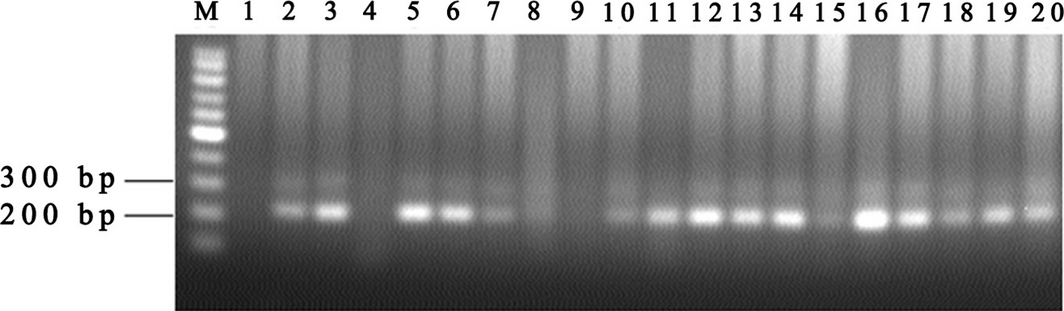
Figure 3. Agarose gel electrophoresis of Toxoplasma gondii infection PCR. M: 100 bp ladder; 1: negative control; 20: positive control; 2, 3, 5, 6, 7, 10, 11, 12, 13, 14, 16, 17, 18, 19: positive PCR products; 4, 8, 9, 15: negative PCR products. |
DNA sequencing and phylogenetic analysis
Four T. gondii amplicons from animals from Sidi Bouzid and four amplicon from Beja were randomly chosen for genetic analysis. They were purified with the ExoASP-IT® (usb) according to the manufacturers instructions and sequenced in both directions using an ABI 3730xl DNA Analyser (96 capillary type). Sequencing reactions were performed in the DNA Engine Tetrad 2 Peltier Thermal Cycler (BIO-RAD) using the ABI BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), following the protocols supplied by the manufacturer. Single-pass sequencing was performed on each template using internal primers (Tg-NP1 and Tg-NP2). The fluorescent-labelled fragments were purified from the unincorporated terminators with the Big Dye XTerminator® Purification Kit (Applied Biosystems). An electrophoresis was performed in an ABI 3730xl DNA Analyser (Applied Biosystems).
The chromatograms were evaluated with ChromasPro software (version 1.7.4). MEGA 5.1 software was used to perform multiple sequence alignments (Tamura et al. 2011), and the sequences were compared with the GenBank database by nucleotide sequence homology search made at the network server of the National Centre for Biotechnology Information (NCBI) with BLAST. A phylogenetic tree of T. gondii was constructed from the ITS1 rDNA gene sequences of our amplicons and those available in GenBank.
Statistical analysis
The infection prevalence percentages were compared using Epi Info 6 (Dean et al. 2011). A chi-square Mantel–Haenszel test was performed. A probability less than 0.05 was used as a threshold for statistical significance (Schwartz 1993).
Results
One hundred and seventy-four animals from Sidi Bouzid belonged to two breeds, namely Barbarine (56.3%) and Queue Fine de l'Ouest (QFO) (43.7%), and were a mixture of lambs and ewe lambs. One hundred and twenty lambs from Beja sheep were Barbarine (75.3%) and cross-breed (24.7%).
Molecular prevalence of T. gondii
The overall prevalence of infection was 31% (54/174) and 32% (48/150) for animals from Sidi Bouzid and Beja, respectively.
There were no statistically significant differences in infection prevalence between the three localities of Sidi Bouzid (Sidi Bouzid North, Centre and South) and between the four age groups (P > 0.05). In Beja region, no significant differences were recorded between animals from different breeds, localities and age groups (P > 0.05) (Table 2).
| Sidi Bouzid | Beja | ||||||
|---|---|---|---|---|---|---|---|
| Parameter | Positive/examined (%) | P value | Parameter | Positive/examined (%) | P value | ||
| Breed | Barbarine | 25/98 (25.5) | 0.07 | Breed | Barbarine | 38/113 (33.6) | 0.46 |
| QFO | 29/76 (38.2) | Cross-breed | 10/37 (27.0) | ||||
| Locality | North Sidi Bouzid | 10/34 (29.4) | 0.3 | Locality | Amdoun | 22/60 (36.6) | 0.4 |
| Centre Sidi Bouzid | 40/118 (33.9) | Beja | 20/63 (31.7) | ||||
| South Sidi Bouzid | 4/22 (18.2) | Thibar and Teboursouk | 6/27 (22.2) | ||||
| Age group (years) | ≤1 | 14/44 (31.8) | 0.8 | Age group (months) | 5 | 13/33 (39.4) | 0.57 |
| 1–3 | 12/37 (32.4) | 6 | 12/48 (25.0) | ||||
| 4–5 | 16/46 (34.8) | 7 | 10/29 (34.5) | ||||
| 6–10 | 12/47 (25.5) | 8–12 | 13/40 (32.5) | ||||
| Overall | 54/174 (31.0) | 48/150 (32.0) | |||||
| QFO, Queue Fine de l'Ouest. | |||||||
Phylogenetic analyses of T. gondii
The BLAST comparison of the partial sequences of the ITS1 rDNA gene (260 bp length) revealed 100% homology between the eight genotypes (GenBank accession no: KT896498). Amplicons from Sidi Bouzid and Beja shared 99.2% homology with the recently reported sequences for the ITS1 rDNA gene of T. gondii from America (KM527503, AY488166, AF252408 and KP999999), Brazil (JF810959, FJ966049 and FJ176233), Norway (KM657806), China (JX456457and AJ628251), Portugal (AY582110) and Germany (EU025025). They shared 98.5% and 97% homology with two Brazilian isolates (JF810935 and GQ160472, respectively). A phylogenetic tree of T. gondii was constructed from the ITS1 rDNA gene sequences of our amplicons and those available in GenBank (Fig. 4).
|
|
|
Figure 4. Phylogenetic relationships based on the partial nucleotide sequences (260 bp) of ITS1 rDNA gene for Toxoplasma gondii detected in Northern (Beja) and Central (Sidi Bouzid) Tunisia (North Africa) with other isolates deposited in GenBank. The host, the country of origin and the GenBank accession numbers are given in parentheses. Sequence obtained in the present study is indicated with a black square. |
Discussion and conclusion
Toxoplasma gondii is a worldwide-distributed parasite that infects all warm-blooded vertebrate species, including human. Acquired human infection is caused by ingestion of either sporulated oocysts from the environment, water or vegetables or bradyzoites in undercooked meat products. Cats are the key in the circle of T. gondii because they are the only domestic hosts that can excrete the environmentally resistant oocysts in their faeces (Dubey 2010).
The molecular detection of T. gondii DNA showed a high prevalence (31% and 32% in Sidi Bouzid and Beja, respectively). These results are different from those previously reported by Gharbi et al. (2013) who investigated three age groups (lambs, yearlings and rams). The T. gondii molecular prevalence in Sidi Bouzid estimated here (31%) was similar to that reported in the same governorate by Gharbi et al. (2013) (25.5%; P = 0.32). In the northern region, our results showed higher molecular prevalence (32%) compared to the 12.7% found by Gharbi et al. (2013) (P = 0.002). Higher molecular prevalence was obtained in ewes’ meat in Tunis City (50%) (Boughattas et al. 2014). A low-molecular prevalence rate of only 5.7% was reported for T. gondii in the hearts’ apexes of 70 sheep slaughtered in households of Central East Tunisia during the Muslim Feast of Sacrifice (Khayeche et al., 2014). This low prevalence found by Khayeche et al. (2014) may be due to the fact that 92.9% of the animals included in the study were aged under 1 year. Results of the current study do not lend support to this hypothesis as we were unable to show differences in prevalence between the different age groups.
On the one hand, molecular prevalence values obtained in some countries were close to results obtained in this trial. In fact, Asgari et al. (2010) showed that the prevalence of T. gondii infection was 37.5% in sheep from Fars province, Southern Iran. Azizi et al. (2014) showed that 38% of sheep in Southwest of Iran were infected with T. gondii. In Morocco, the DNA of T. gondii was identified by nested PCR in 38% of studied sheep (Azizi et al. 2014).
On the other hand, many studies showed higher or lower prevalence values of the infection. For example, in Ireland, the presence of T. gondii DNA was detected in diaphragm samples from 3.6% (3/83) sheep (Halová et al. 2013), and higher prevalence values (67.7%) were found in Egypt (Ghoneim et al. 2010).
In the present study, only 0.5 g meat samples were analysed which could induce an under-estimation of T. gondii infection prevalence, the ability of gene amplification by PCR allowed detection of small amounts of DNA. This suggests that PCR is a sensitive method for the diagnosis of toxoplasmosis (Wastling et al. 1993). Because of high contamination risks of the two-step nested PCR, drastic measures to prevent contamination were observed along all the PCR runs.
No significant differences were found between animals from different breeds, localities within district and age groups. This result is in concordance with the result obtained by Gharbi et al. (2013). This should be due to the presence of the same risk factors in the studied animal population. In Bangladesh, authors found that the age was not a significant risk factor in T. gondii infection (Rahman et al. 2014). However, Boughattas et al. (2014) found a difference in prevalence values between ages. Hecker et al. (2013) found a significantly higher proportion of T. gondii seropositive animals in females and older sheep. Some authors established an association between T. gondii infection and age (Guimarães et al. 2013). Other ones found that the most significant factors associated with T. gondii infection in sheep were age and gender (Cosendey-KezenLeite et al. 2014).
Even if climate plays an important role in the conservation of oocysts of T. gondii, because occurrence locations of T. gondii infection are related to higher temperature, lower precipitation and lower altitude compared to the absence locations of T. gondii (Kantzoura et al. 2013), this study did not reveal a difference in infection prevalence between the two regions. Although some authors found differences in prevalence values according to age and region, results of this survey did not bring out these differences. This may be due to the sampling. In fact, the small number of animals did not detect statistically significant differences related to some risk factors.
Infections caused by T. gondii were confirmed in ewes using a nested PCR followed by sequencing of the ITS1 region of the parasites’ rDNAs. All sequences shared 97–99% identity with sequences from other strains present in GenBank. In fact, the two T. gondii sequences were identical to many ones, namely America, Norway, China, Brazil and Germany, and had a high genetic homology with sequence from Brazil. Toxoplasma gondii from sheep showed high genetic diversity. In Tunisia, sheep meat is highly infected by T. gondii representing though a major risk for the consumer especially for pregnant women who should be informed about this risk and the specific prevention majors that they should practice (avoid manipulation of lamb meat and consumption of undercooked meat). Further studies are needed to improve our knowledge on different genotypes of T. gondii that infect Tunisian sheep population.
Acknowledgements
The study was financially supported by ‘Laboratoire d'épidémiologie d’infections enzootiques des herbivores en Tunisie: application à la lutte' (Ministère de l'enseignement supérieur et de la recherche scientifique, Tunisia). This work was partly supported by ICARDA-led research program CRP dryland systems.
Source of funding
The work was funded by the laboratory Epidemiology of endemic infections of herbivores in Tunisia: application to the control. Ministry of Higher Education, Scientific Research and Information Technology and Communication, Tunisia. This work was partly supported by ICARDA-led research program Consultative Group on International Agricultural Research Programme (CRP) dryland systems.
Conflicts of interest
The authors declare no conflicts of interest in relation to this work.
Contributions
M Rouatbi, M Gharbi and M Rekik conceived and designed the experiments. M Rouatbi, S Amairia and MR Rjeibi performed the experiments. M Rouatbi, Y Amdouni and S Sammoudi involved in the collection of samples. M Rekik, M Gharbi and M Rouatbi wrote the manuscript.
References
- Climate data.org. Available at: http://fr.climate-data.org/ (Accessed 16 November 2015).
- Dateandtime.info. Available at: http://dateandtime.info/ (Accessed 16 November 2015).
- Akkari H., Gharbi M. & Darghouth M.A. (2011) Infestation of tracer lambs by Fasciola hepatica in Tunisia: determining periods for strategic anthelmintic treatments. Revue Scientifique et Technique-OIE30, 917–929.
- Akkari H., Gharbi M. & Darghouth M.A. (2012) Dynamics of infestation of tracers lambs by gastrointestinal helminths under a traditional management system in the North of Tunisia. Parasite19, 407–415.
- Alazemi M.S. (2014) Prevalence of anti-Toxoplasma gondii antibodies in aborted ewes in Kuwait. Journal of the Egyptian Society of Parasitology44, 393–396.
- Amairia S., Rouatbi M., Rjeibi M.R., Nouasri H., Sassi L., Mhadhbi M. & Gharbi M. (2016) Molecular prevalence of Toxoplasma gondii DNA in goats’ milk and seroprevalence in Northwest Tunisia. Veterinary Medicine and Science2, 154–160. doi:10.1002/vms3.29.
- Asgari Q., Sarnevesht J., Kalantari M., Adnani Sadat J., Motazedian M.H. & Sarkari B. (2010) Molecular survey of Toxoplasma infection in sheep and goat from Fars province, Southern Iran. Tropical Animal Health and Production43, 389–395.
- Azizi H., Shiran B., Boroujeni A.B. & Jafari M. (2014) Molecular Survey of Toxoplasma gondii in Sheep, Cattle and Meat Products in Chaharmahal va Bakhtiari Province, Southwest of Iran. Iranian Journal of Parasitology9, 429–434.
- Ben Salem H., Lassoued N. & Rekik M. (2011) Merits of the fat-tailed Barbarine sheep raised in different production systems in Tunisia: digestive, productive and reproductive characteristics. Tropical Animal Health and Production43, 1357–1370.
- Bezerra M.J., Cruz J.A., Kung E.S., Albuquerque P.P., Kim P.C., Moraes E.P.et al. (2014) Detection of Toxoplasma gondii DNA in fresh and frozen semen from rams in Brazil. Reproduction in Domestic Animals49, 753–755.
- Boughattas S., Ayari K., Sa T., Aoun K. & Bouratbine A. (2014) Survey of the Parasite Toxoplasma gondii in Human Consumed Ovine Meat in Tunis City. PLoS ONE. doi:10.1371/journal.pone.0085044.
- Bouratbine A., Siala E., Chahed M.K., Aoun K. & Ben Ismail R. (2001) Sero-epidemiologic profile of toxoplasmosis in northern Tunisia. Parasite.8, 61–66.
- Buxton D. & Finlayson J. (1986) Experimental infection of pregnant sheep with Toxoplasma gondii: pathological and immunological observations on the placenta and foetus. Journal of Comparative Pathology96, 319–333.
- Buxton D., Thomson K., Maley S., Wright S. & Bos H.J. (1991) Vaccination of sheep with a live incomplete strain (S48) of Toxoplasma gondii and their immunity to challenge when pregnant. The Veterinary Record129, 89–93.
- Cosendey-KezenLeite R.I., De Oliveira F.C., Frazão-Teixeira E., Dubey J.P., De Souza G.N., Ferreira A.M. & Lilenbaum W. (2014) Occurrence and risk factors associated to Toxoplasma gondii infection in sheep from Rio de Janeiro, Brazil. Tropical Animal Health and Production46, 1463–1466.
- Dean A.G., Arner T.G., Sunki G.G., Friedman R., Lantinga M., Sangam S.et al. (2011) Epi Info ™ A Database and Statistics Program for Public Health Professionals. CDC: Atlanta, GA, USA.
- Długońska H. (2014) Toxoplasma gondii and the host cells. Annals of Parasitology60, 83–88.
- Dubey J.P. (2010) Toxoplasmosis of Animals and Humans. 2nd edn. CRC PressBoca Raton, Florida.
- Dubey J.P., Casey S.J., Zajac A.M., Wildeus S.A., Lindsay D.S., Verma S.K.et al. (2014) Isolation and genetic characterization of Toxoplasma gondii from alpaca (Vicugna pacos) and sheep (Ovis aries). Tropical Animal Health and Production46, 1503–1507.
- Dunn D., Wallon M., Peyron F., Petersen E., Peckham C. & Gilbert R. (1999) Mother-to-child transmission of toxoplasmosis: risk estimates for clinical counselling. Lancet353, 1829–1833.
- Flatt A. & Shetty N. (2013) Seroprevalence and risk factors for toxoplasmosis among antenatal women in London: a re-examination of risk in an ethnically diverse population. The European Journal of Public Health23, 648–652. doi:10.1093/eurpub/cks075.
- Gelaye W., Kebede T. & Hailu A. (2015) High prevalence of anti-toxoplasma antibodies and absence of Toxoplasma gondii infection risk factors among pregnant women attending routine antenatal care in two Hospitals of Addis Ababa, Ethiopia. International Journal of Infectious Diseases34, 41–45. doi:10.1016/j.ijid.2015.03.005.
- Gharbi M., Zribi L., Jedidi M., Chakkhari H., Hamdi S., R'hayem S.et al. (2013) Prévalence d'infection des ovins par Toxoplasma gondii en Tunisie. Prevalence of Toxoplasma gondii infection in Tunisian sheep. Bulletin de la Societe de Pathologie Exotique106, 184–187.
- Ghoneim N.H., Shalaby S.I., Hassanain N.A., Zeedan G.S., Soliman Y.A. & Abdalhamed A.M. (2010) Comparative study between serological and molecular methods for diagnosis of toxoplasmosis in women and small ruminants in Egypt. Foodborne Pathogens and Disease7, 17–22.
- Guimarães L.A., Bezerra R.A., Rocha Dde S. & Albuquerque G.R. (2013) Prevalence and risk factors associated with anti-Toxoplasma gondii antibodies in sheep from Bahia state, Brazil. Revista Brasileira de Parasitologia Veterinária22, 220–224.
- Halová D., Mulcahy G., Rafter P., Turčeková L., Grant T. & De Waal T. (2013) Toxoplasma gondii in Ireland: seroprevalence and novel molecular detection method in sheep, pigs, deer and chickens. Zoonoses Public Health60, 168–173.
- Hecker Y.P., Moore D.P., Manazza J.A., Unzaga J.M., Späth E.J., Pardini L.L.et al. (2013) First report of seroprevalence of Toxoplasma gondii and Neospora caninum in dairy sheep from Humid Pampa, Argentina. Tropical Animal Health and Production45, 1645–1647.
- Hurtado A., Aduriz G., Moreno B., Barandika J. & García-Pérez A.L. (2001) Single tube nested PCR for the detection of Toxoplasma gondii in fetal tissues from naturally aborted ewes. Veterinary Parasitology102, 17–27.
- Johnston W.S. (1988) An investigation into toxoplasmosis as a cause of barrenness in ewes. Veterinary Record122, 283–284.
- Jones J.L., Lopez A., Wilson M., Schulkin J. & Gibbs R. (2001) Congenital toxoplasmosis: a review. Obstetrical & Gynecological Survey56, 296–305.
- Kantzoura V., Diakou A., Kouam M.K., Feidas H., Theodoropoulou H. & Theodoropoulos G. (2013) Seroprevalence and risk factors associated with zoonotic parasitic infections in small ruminants in the Greek temperate environment. Parasitology International62, 554–560.
- Khayeche M., Mhadhbi M., Gharbi M., Nasfi I. & Darghouth M.A. (2014) Détection de l'infection par Toxoplasma gondii des ovins abattus dans le gouvernorat de Sousse (centre-est de la Tunisie) à l'occasion de la fête musulmane du sacrifice (Aïd Al-Adha) et analyse des facteurs de risque. Detection of Toxoplasma gondii infection of sheep slaughtered in the governorate of Sousse on the occasion of the Muslim sacrifice feast (Eid Al-Adha) and analysis of risk factors. Bulletin de la Societe de Pathologie Exotique107, 60–63.
- Lahmar S., Trifi M., Naceur S.B., Bouchhima T., Lahouar N., Lamouchi I.et al. (2012) Cystic echinococcosis in slaughtered domestic ruminants from Tunisia. Journal of Helminthology87, 318–325.
- Leblebicier A. & Yildiz K. (2014) Seroprevalence of Toxoplasma gondii in Sheep in Silopi District by Using Indirect Fluorescent Antibody Test (IFAT). Türkiye Parazitolojii Dergisi38, 1–4.
- Ministry of Agriculture. (2014) Official statistics of the Tunisian ministry of agriculture, Survey on agricultural season (Livestock), General Administration of Studies and Agricultural Development, Department of Statistics and Economic circumstance agricultural.
- Opsteegh M., Teunis P., Mensink M., Zuchner L., Titilincu A., Langelaar M. & van der Giessen J. (2010) Evaluation of ELISA test characteristics and estimation of Toxoplasma gondii seroprevalence in Dutch sheep using mixture models. Preventive Veterinary Medicine96, 232–240.
- Perry B., Grieve A., Mogollon J. & de Galvis A. (1978) Serological study of ovine toxoplasmosis in Colombia: prevalence of haemagglutinating antibodies to toxoplasma in sheep. The Veterinary Record103, 584–585. doi:10.1136/vr.103.26-27.584.
- Rahman M., Azad M.T., Nahar L., Rouf S.M., Ohya K., Chiou S.P.et al. (2014) Age-specificity of Toxoplasma gondii seroprevalence in sheep, goats and cattle on subsistence farms in Bangladesh. Journal of Veterinary Medical Science76, 1257–1259.
- Samra N.A., McCrindle C.M.E., Penzhorn B.L. & Cenci-Goga B. (2007) Seroprevalence of toxoplasmosis in sheep in South Africa. Journal of the South African Veterinary Association78, 116–120.
- de Santana Rocha D., de Sousa Moura’ R.L., Maciel B.M., Guimarães L.A., O'dwyer H.N.S., Munhoz A.D. & Albuquerque G.R. (2015) Detection of Toxoplasma gondii DNA in naturally infected sheeps milk. Genetics and Molecular Research14, 8658–8662. doi:10.4238/2015.July.31.14.
- Schwartz D. (1993) Méthodes Statistiques à L'usage des Médecins et des Biologistes, 3ème éd. Flammarion: Paris, France.
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M. & Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution28, 2731–2739.
- Torgerson P.R., Devleesschauwer B., Praet N., Speybroeck N., Willingham A.L., Kasuga F.et al. (2015) World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: a Data Synthesis. PLoS Medicine12, doi:10.1371/journal.pmed.1001920.
- Van der Puije W.N., Bosompem K.M., Canacoo E.A., Wastling J.M. & Akanmori B.D. (2000) The prevalence of anti-Toxoplasma gondii antibodies in Ghanaian sheep and goats. Acta Tropica76, 21–26.
- Wang Y., Wang G., Zhang D., Yin H. & Wang M. (2013) Detection of Acute Toxoplasmosis in Pigs Using Loop-Mediated Isothermal Amplification and Quantitative PCR. Korean Journal of Parasitology51, 573–577. doi:10.3347/kjp.2013.51.5.573.
- Wang Y., Tian R.M., Gao Z.M., Bougouffa S. & Qian P.Y. (2014) Optimal Eukaryotic 18S and Universal 16S/18S Ribosomal RNA Primers and Their Application in a Study of Symbiosis. PLoS ONE9. doi: 10.1371/journal.pone.0090053.
- Wastling J.M., Nicoll S. & Buxton D. (1993) Comparison of two gene amplification methods for the detection of Toxoplasma gondii in experimentally infected sheep. Journal of Medical Microbiology38, 360–365.
Document information
Published on 09/06/17
Submitted on 09/06/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?