(Created page with " ==Abstract== Interest in the composition of the intestinal microbiota and possibilities of its therapeutic modifications has soared over the last decade and more detailed k...") |
m (Scipediacontent moved page Draft Content 984459227 to Schmitz Suchodolski 2016a) |
(No difference)
| |
Latest revision as of 10:27, 9 June 2017
Abstract
Interest in the composition of the intestinal microbiota and possibilities of its therapeutic modifications has soared over the last decade and more detailed knowledge specific to the canine microbiota at different mucosal sites including the gut is available. Probiotics, prebiotics or their combination (synbiotics) are a way of modifying the intestinal microbiota and exert effects on the host immune response. Probiotics are proposed to exert their beneficial effects through various pathways, for example production of antimicrobial peptides, enhancing growth of favourable endogenous microorganisms, competition for epithelial colonisation sites and immune-modulatory functions. Despite widespread use of pro-, pre- and synbiotics, scientific evidence of their beneficial effects in different conditions of the dog is scarce. Specific effects of different strains, their combination or their potential side-effects have not been evaluated sufficiently. In some instances, in vitro results have been promising, but could not be transferred consistently into in vivo situations. Specific canine gastrointestinal (GI) diseases or conditions where probiotics would be beneficial, their most appropriate dosage and application have not been assessed extensively. This review summarises the current knowledge of the intestinal microbiome composition in the dog and evaluates the evidence for probiotic use in canine GI diseases to date. It wishes to provide veterinarians with evidence-based information on when and why these products could be useful in preventing or treating canine GI conditions. It also outlines knowledge about safety and approval of commercial probiotic products, and the potential use of faecal microbial transplantation, as they are related to the topic of probiotic usage.
Introduction
Microorganisms are found abundantly in association with mammalian hosts; in fact, the number of microbial cells is around 10 times that of host cells (Gibson & Roberfroid 1995), consisting of around 1000 times more microbial genes (The Human Microbiome Project; www.http://hmpdacc.org). The concept of the microbiome was first suggested by Joshua Lederberg, who coined the term ‘microbiome’ to ‘signify the ecological community of commensal, symbiotic and pathogenic microorganisms that literally share our body space’ (Lederberg & McCray 2001). It is generally accepted that the term ‘microbiota’ (in the past also referred to as microflora) is used to describe bacterial communities on mucosal surfaces (with or without luminal microorganisms) or on other body sites (e.g. skin). Overlapping, but distinct, the term ‘microbiome’ is nowadays used to refer to the entire genetic mass (‘genome’) of microorganisms. It is mostly, but incorrectly, used to describe bacterial genomic communities. One should probably refer specifically to the ‘bacteriome’, ‘virome’(Mansfield 2015) or ‘mycobiome’ (Foster et al. 2013), respectively, and use the term microbiome globally for all microorganisms combined (ten Oever & Netea 2014). These microorganisms include eucaryotes, archaea, bacteria, viruses and fungi. Living in such close contact, they usually are not harmful to the host; but considered beneficial in most cases. For example the intestinal microbiota supplies short-chain fatty acids (SCFA) as nutrients for colonocytes (Cummings 1981; Rérat et al. 1987; Cummings & Macfarlane 1991).
In recent years, more and more research has focused on characterising microbiota at different body sites in people, leading to the Human Microbiome Project (The NIH HMP Working Group 2009); and considerable progress has also been made in defining and understanding microbial communities in small animals, especially as the availability of large-scale genomic sequencing techniques.
Naturally, the detection of differences in microbiota characteristics or the microbiome composition between healthy and diseased subjects has led to the conclusion that modifying these microbial communities might have a beneficial effect on host health in certain circumstances. This is where the application of pre- or probiotics or their combination (so called synbiotics) has been the focus of much attention; especially in human and canine intestinal diseases like inflammatory bowel disease (IBD) (Sauter et al. 2006; Ghouri et al. 2014; Rossi et al. 2014; Schmitz et al. 2014, 2015a; Saez-Lara et al. 2015).
As probiotics are not usually defined as drugs, they do not have to undergo any process proving their efficacy in applications, diseases or even target species. Hence, many medical claims have been made regarding their beneficial effects, both in humans and in animals. This review focuses on what is known about the definitions, mechanisms of action and efficacy of probiotics in different GI diseases in dogs, and summarises these findings so that the reader can make a better judgement about when and how to use probiotics in small animal gastroenterology.
Composition of the gastrointestinal microbial communities in dogs
The gastrointestinal microbiome in healthy dogs
Recently, high-throughput DNA sequencing techniques have improved microbial identification in small animals. Mostly, these techniques have been applied to describe the phylogenetic structure and functional capacity of the GI microbiome (Handl et al. 2011; Swanson et al. 2011; Hooda et al. 2012). Some studies have included mucosal samples or intestinal content of different segments of the GI tract (Suchodolski et al. 2009, 2010, Suchodolski et al. 2012a), but most have focused on analysis of faecal samples, as these are easier to obtain (Garcia-Mazcorro et al. 2011; Handl et al. 2011, 2013; Swanson et al. 2011; Suchodolski et al. 2012b; Honneffer et al. 2014; Minamoto et al. 2014a). This is important when comparing studies with each other, as bacterial populations have been shown to differ between different substrates and intestinal sites (Momozawa et al. 2011).
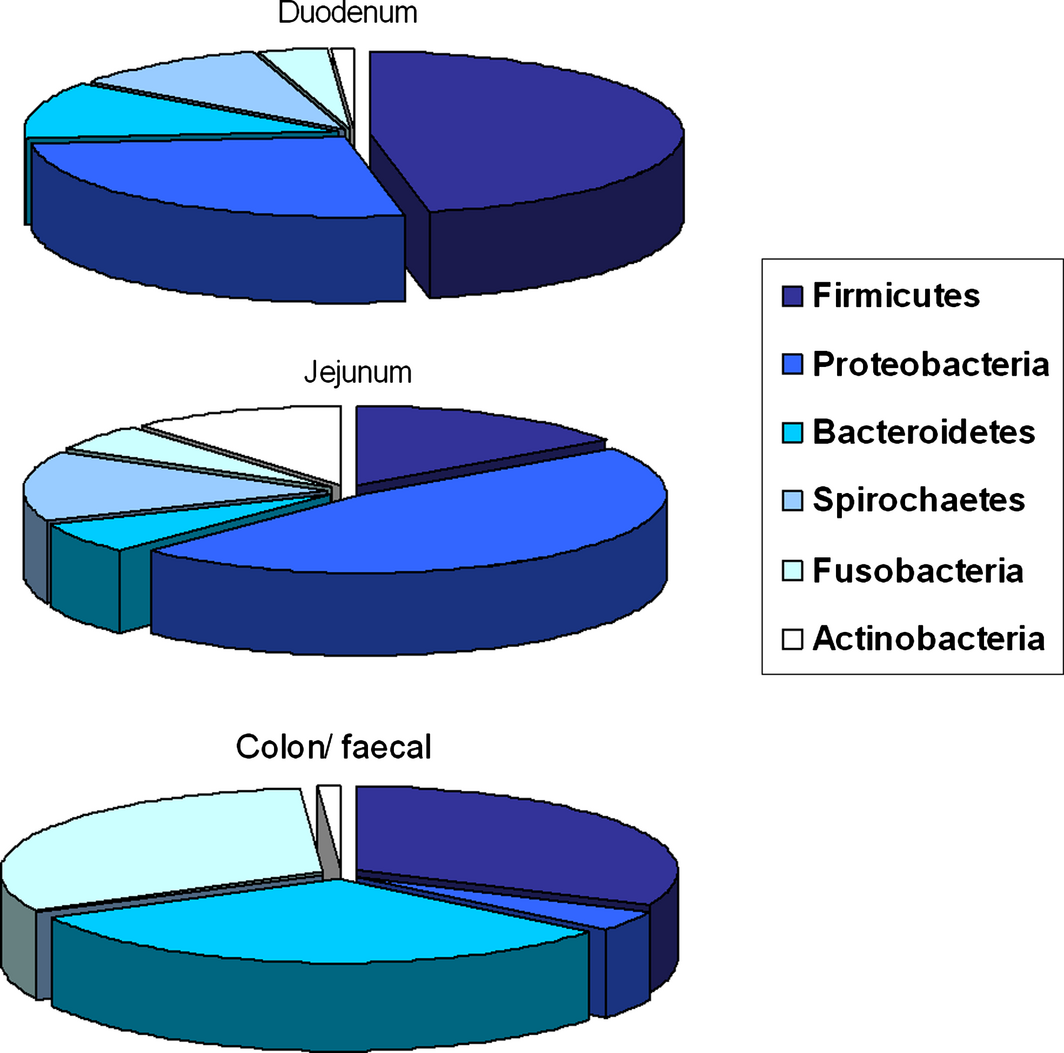
In general, bacterial number and diversity increase gradually along the GI tract (Suchodolski et al. 2005) (Fig. 1). In the healthy canine stomach, total bacterial load is comparably low (105 log10 16S rRNA copy numbers), and mostly belong to Proteobacteria (99.6% of obtained gene sequences) with only few Firmicutes (0.3%) (Garcia-Mazcorro et al. 2012). The predominant species are Helicobacter and Lactobacillus spp. The healthy canine duodenal microbial community as reported by one study to consist of six primary phyla: Firmicutes (46.4% of obtained 16S rRNA gene sequences), Proteobacteria (26.6%), Bacteroidetes (11.2%), Spirochaetes (10.3%) Fusobacteria (3.6%) and Actinobacteria (1%) (Xenoulis et al. 2008). Another study evaluated the microbiota in the jejunum of healthy dogs, and identified Proteobacteria as the most abundant (46%), followed by Firmicutes (15%), Actinobacteria (11.2), Spirochaetes (14.2), Bacteroidetes (6.2%) and Fusobacteria (5.4%) (Suchodolski et al. 2009) (Fig. 1). Duodenal and jejunal ingesta samples contained 22% and 10% of Lactobacillales respectively (Xenoulis et al. 2008). Suchodolski et al. (2009) also identified four additional phyla in the jejunum that were not reported in dogs previously: Tenericutes, Cyanobacteria, Verrucomicrobia and Chloroflex, which were all present in low frequency (<0.1%) (Suchodolski et al. 2009). The ileal ingesta from healthy research dogs predominantly contained Fusobacteria, Firmicutes and Bacteroidetes (Suchodolski et al. 2008). This is significantly different to the composition of the duodenal and jejunal microbiome composition, as especially the orders of Fusobacteriales (30%), and Clostridiales (22%) with both Clostridium clusters XI and XIVa were predominant in the healthy ileum. Also in contrast to duodenal and jejunal samples, ileal contents had much lower proportions of Lactobacillus spp. (1.4%). Whether these variations are true qualitative and quantitative differences is hard to assess, as some of them – especially between different studies – are likely due to variations in the different DNA extraction methods, differences in amplification primers, and sequencing platforms used (e.g. 454-pyrosequencing vs. 16S rRNA clone libraries). Colon samples showed the co-dominant phyla of Fusobacteria, Bacteroidetes and Firmicutes (around 30% each) in healthy dogs (Suchodolski et al. 2008). The presence of Fusobacteria (108 cfu/ml of intestinal content) also has been demonstrated using culture-based techniques (Davis et al. 1977). The next abundant order present was Clostridiales (18%), with Clostridum cluster XIVa being the predominant member (50%) (Suchodolski et al. 2008). This cluster includes Eubacterium, Roseburia and Ruminococcus spp., which are dietary fibre fermenters. Proteobacteria, including E. coli-like organisms, were present in low proportions (1.4%), whereas Lactobacillales where present at levels similar to the jejunum (10%) (Suchodolski et al. 2008). Results of the analyses of the canine faecal microbiome indicated a predominance of the phyla Fusobacteria (24–40%), Bacteroidetes (32–34%), Firmicutes (15–28%), Proteobacteria (5–6%) and Actinobacteria (0.8–1.4%) (Xenoulis et al. 2008; Suchodolski et al. 2009; Middelbos et al. 2010; Handl et al. 2011; Swanson et al. 2011; Garcia-Mazcorro et al. 2012) (Fig. 1).
|
|
|
Figure 1. Distribution of typical bacterial phyla within different compartments of the intestinal tract in dogs. |
The effect of dietary intervention on the canine gastrointestinal microbial composition
Even though some studies have provided evidence that the administration of prebiotics/fibre in the diet has the ability to manipulate the GI microbiota of dogs (Spears et al. 2005; Beloshapka et al. 2013; Panasevich et al. 2014), there are some limitations. Firstly, traditional plating techniques or qPCR to quantify a limited number of bacteria (e.g. Lactobacillus sp., Bifidobacterium sp., Clostridia, E. coli) were mostly used (Spears et al. 2005; Strompfová et al. 2012a). Second, most studies were performed in healthy dogs, which make inferring results to diseased dogs or animals susceptible to GI disturbances (e.g. weanlings and geriatrics) difficult. Third, faecal samples were analysed rather than mucosal biopsies or ingesta (Spears et al. 2005; Verlinden et al. 2006; Hang et al. 2012; Strompfová et al. 2012b). Lastly, a wide range of prebiotic products and dosages have been administered, making comparisons between studies difficult. The authors of this review recently analysed the faecal microbiome of dogs with food-responsive chronic enteropathy (CE) before and after 6 weeks of dietary intervention and compared these to the faecal microbiome of healthy dogs before and after they were switched to the same diet used in the diseased dogs (a hydrolysed protein diet), and could not detect a significant effect on microbial composition or diversity attributed to the dietary change (Schmitz et al., unpublished data). More research using molecular sequencing techniques is clearly needed to examine the effect of dietary interventions in healthy dogs and dogs with GI disease.
The canine gastrointestinal microbial composition in disease
The invasion and/or colonisation of the GI tract with specific pathogens may profoundly disturb the integrity of the intestinal epithelial barrier (Viswanathan et al. 2009). Several potential GI pathogens are recognised in dogs, including Clostridium perfringens, Salmonella spp. and E. coli (Marks et al. 2002). However, most of those are also recognised commensals and have been isolated at similar frequencies from dogs with and without signs of GI disease (Marks & Kather 2003; Unterer et al. 2014; Busch et al. 2015). Therefore, the cause and effect relationship between those organisms and GI disease needs to be interpreted with caution.
Non-specific alterations of the GI microbiota have been regarded as a pivotal factor for the development of acute or chronic GI disease. Several studies have attempted to characterise the faecal microbial composition in diarrhoeic dogs. In acute diarrhoea, large-scale changes were observed, both with culture and sequencing techniques. This included increased abundance of Clostridium spp. (especially C. perfringens), E. coli, Lactobacillus and Enterococcus spp. with concurrent reductions of those bacterial groups that make up the majority of the normal colonic microbiota, such as Faecalibacterium, Ruminococcaceae and Blautia spp. (Bell et al. 2008; Minamoto et al. 2014b; Guard et al. 2015). In chronic diarrhoea, significantly higher counts of Bacteroides sp. were found using fluorescent in situ hybridisation analysis (Jia et al. 2010). In a study evaluating a large group of dogs with chronic diarrhoea using qPCR assays, diseased dogs had significantly decreased abundances of Fusobacteria, Ruminococcaceae, Blautia spp. and Faecalibacterium spp. and significantly increased abundances of Bifidobacterium spp., Lactobacillus spp. and E. coli compared to healthy dogs (Minamoto et al. 2014b).
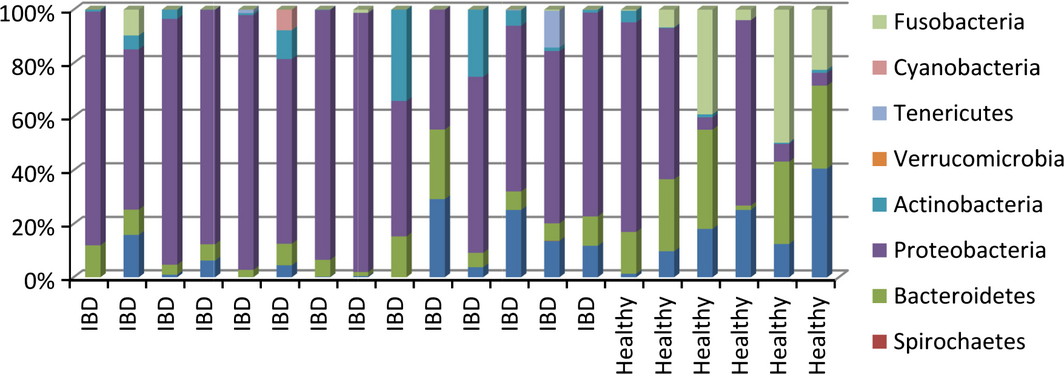
In samples from dogs with IBD, microbiome changes similar to the ones observed in people with IBD were detected. A significantly reduced species richness and higher proportion of Enterobacteriaceae were observed in duodenal brush samples from dogs with IBD compared with healthy dogs (Xenoulis et al. 2008). Additionally, in duodenal mucosal biopsies, a higher abundance of Proteobacteria and lower amount of Clostridia were found in IBD compared with healthy dogs (Suchodolski et al. 2010) (Fig. 2). The analysis of faecal samples from dogs with IBD revealed dysbiosis, with significantly lower bacterial diversity, an increase in Gammaproteobacteria (i.e. E. coli) and decreases in Erysipelotrichia, Clostridia and Bacteroidia (Minamoto et al. 2014a,b).
|
|
|
Figure 2. Distribution of bacterial phyla in the duodenum of 14 dogs with inflammatory bowel disease (IBD) and six healthy dogs (based on: Suchodolski et al. 2012a, b). |
Whether these changes are partially a cause or a result of the aberrant immune reactions seen in the GI tract in IBD remains a matter of debate, both in people and in dogs. However, it is now suspected that these bacterial changes are associated with altered metabolic functions of the microbiota (e.g. decrease in SCFA concentrations, altered amino acid metabolism, changed in redox equilibrium, altered bile acid metabolism), and are therefore potentially exacerbating the inflammatory state of the host (Hall 2011; Minamoto et al. 2014a; Guard et al. 2015).
Definition of probiotics, prebiotics and synbiotics
Probiotics are most frequently defined as live microorganisms, which when consumed in adequate amounts confer a health benefit on the host (FAO/WHO, 2002). However, in a lot of circumstances, their health benefits are not strictly proven for a given disease, application or host organism, but the term probiotics is still used. It would be more appropriate to describe probiotics in small animals as live microorganisms given with the intention of improving host health. They include exogenous and indigenous bacterial species that interact with various cellular components within the host (see below).
Prebiotics are defined as selectively fermented ingredients that result in specific changes in the composition and/or activity of the gastrointestinal microbiota, thus also being a benefit to the host organism (Gibson et al. 2010; Roberfroid et al. 2010). Usually, prebiotics are fibre compounds of different length that pass undigested through the gastrointestinal tract. These include disaccharides (lactulose, tagatose), oligo- or polysaccharides [fructo-oligosaccharides (FOS), mannan oligosaccharides (MOS) xylooligosaccharides, polydextrose, galacto oligosaccharides] or long-chain prebiotics like inulin (Hughes & Rowland 2001; Ogué-Bon et al. 2010; Roberfroid et al. 2010; Koh et al. 2013).
Finally, synbiotics are preparations combining both probiotics and prebiotics. This concept was first introduced as ‘mixtures of probiotics and prebiotics that beneficially affect the host by improving the survival and implantation of live microbial dietary supplements in the gastrointestinal tract, by selectively stimulating the growth and/or by activating the metabolism of one or a limited number of health-promoting bacteria, thus improving host welfare’ (Gibson & Roberfroid 1995). The Food and Agriculture Organization of the United Nations (FAO) recommends that the term synbiotic should be used only if the net health benefit observed is synergistic.
Mechanisms of probiotic action
Probiotics can enhance mucosal health by several proposed mechanisms, including displacement of intestinal pathogens (Lee et al. 2003), production of antimicrobial substances (Jones & Versalovic 2009), enhancement of immune responses (Pagnini et al. 2010), and/or up-regulation of various metabolites (Soo et al. 2008).
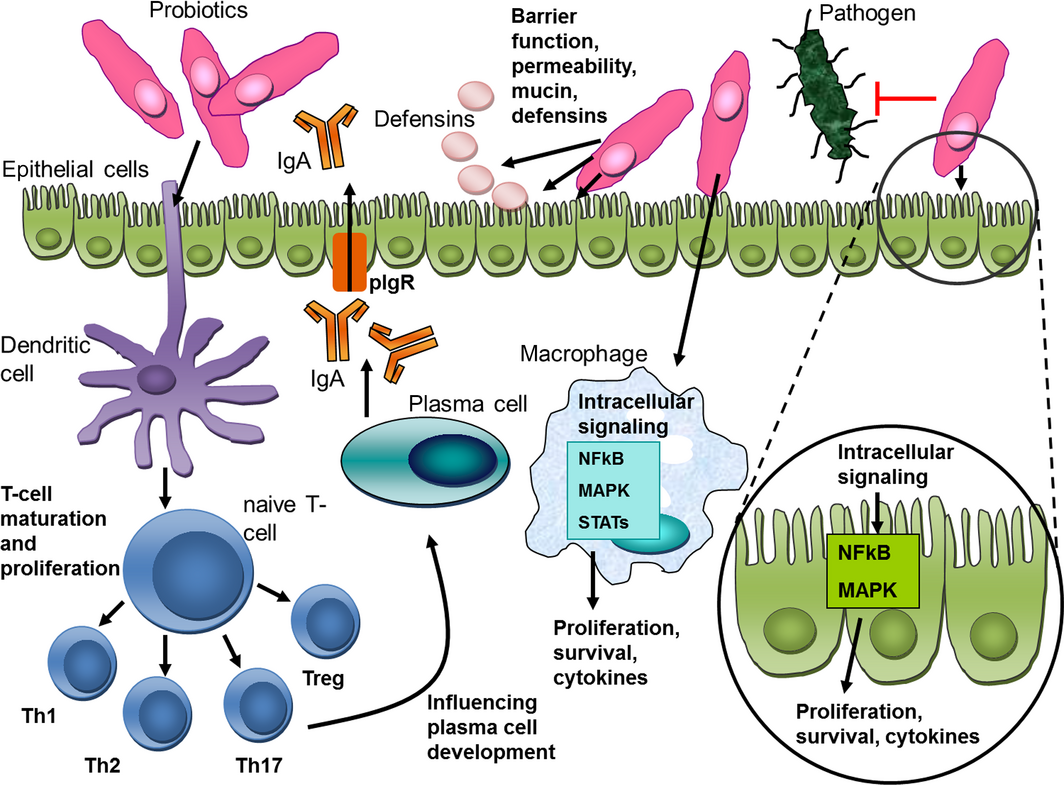
Probiotics can compete with potential pathogens by interfering with their adherence to the intestinal mucosa or by induction of mucus/mucin production (Collado et al. 2007a). These mechanisms are thought to be strain specific, with some strains having increased adherence capabilities (e.g. L. rhamnosus GG = LGG), and some strains being able to increase the adherence of pathogens to intestinal mucus (Collado et al. 2007b). In addition, probiotic bacteria can produce various antimicrobial substances, for example fatty acids, lactic acid and acetic acid (Saarela et al. 2000). Some Lactobacillus spp. can decrease toxin gene expression and production by Salmonella, E.coli or C. perfringens in vitro (Medellin-Peña et al. 2007; Allaart et al. 2011; Bayoumi & Griffiths 2012) or inactivate toxins by production of proteases ex vivo (Castagliuolo et al. 1999). Immune modulation of the host organism – especially intestinal epithelial cells (IECs)– might occur through microbial cell wall components, their metabolites or DNA (Oelschlaeger 2010; Thomas & Versalovic 2010). The effects (again mostly shown in vitro, but also in some animal models of inflammation) include maintenance and fortification of tight junctions, prolonging the survival of IECs and induction of IgA and β-defensin production (Oelschlaeger 2010; Thomas & Versalovic 2010).
Intact, viable bacteria may be essential for probiotic effects, or these effects could be mediated by a cell wall component or structurally diverse secreted molecules, e.g. peptides, lipopeptides, lipopolysaccharides (LPS), DNA, RNA (Lauková et al. 2004). Several mechanistic studies show that key biological signalling pathways like nuclear factor kappa B (NFκB), mitogen-activated protein (MAP) kinases, Akt/phosphatidyl inositol-3 kinase (PI3K) and peroxisome proliferated activator of transcription receptor gamma (PPARγ) are targets for probiotics or their products both in vitro and in vivo (Thomas & Versalovic 2010) (Fig. 3). These pathways can be modified in different ways by individual probiotic strains. This is clearly a strain-specific effect, as even bacterial strains of the same species can alter cellular responses differentially. For example, Lactobacillus reuteri ATCC PTA 6475 can inhibit LPS-induced tumour-necrosis factor alpha (TNFα) production from myeloid cells in vitro through suppression of the activator protein-1 (AP-1) pathway, whereas another L. reuteri strain, DSM 17938, does not inhibit LPS-induced TNFα production (Lin et al. 2009). The details on cellular interactions of specific probiotic strains have been summarised in several reviews (Oelschlaeger 2010; Thomas & Versalovic 2010; Fijan 2014; Vitetta et al. 2014) (figure 3).
|
|
|
Figure 3. Proposed mechanisms of action of probiotics (modified from Thomas & Versalovic 2010). |
Effects of probiotics on intestinal and overall health have mostly been studied in humans and rodent models of human disease (Culligan et al. 2009); much more limited data are available for veterinary species (Callaway et al. 2008). Although probiotics and prebiotics are administered to dogs with increasing frequency, only few investigations have evaluated the complex interplay of probiotics with small animal host cells, immune function or their effect on the intestinal microbial composition (Garcia-Mazcorro et al. 2011). Most of these in vivo studies in dogs were rather crude ex vivo experiments (Sauter et al. 2005; Schmitz et al. 2013, 2014). In addition, most investigations specific for companion animals have only studied the effects of selected probiotic strains or probiotic mixtures on the microbiome or other target effects, for example cytokines (Sauter et al. 2005; Schmitz et al. 2013, 2014).
Microbial organisms commercially used as probiotics in dogs in Europe
To date, four bacterial strains/products have been examined by the European Food Safety Authority (EFSA) for their safety and efficacy as probiotics or feed additives in dogs. This includes two Enterococcus faecium strains (E. faecium NCIMB 10415 E1705, E. faecium NCIMB 10415 E1707), Lactobacillus acidophilus DSM 13241 25 and Bifidobacterium sp. animalis.
Both products containing an E. faecium strain had already been approved for the use in farm animals at the time approval for small animals was sought (2004). For one of these strains, EFSAs conclusion was that enough information was provided to consider it safe for the use in dogs and for humans having contact with treated dogs (E. faecium NCIMB 10415 E1707). The other E. faecium, strain NCIMB 10415 E1705, was considered unlikely to represent a hazard for the target species even when supplied in overdose. It was shown to not favour the growth and shedding of haemolytic and non-haemolytic E.coli in dogs (and cats).
For the product containing Lactobacillus acidophilus DSM 13241 25, EFSA did not establish a safety concern, as the strain was sensitive to medically relevant antibiotics, with the exception of ciprofloxacin. As no data on the effect of this probiotic on shedding of intestinal pathogens in the dog were provided, and it was considered a potential respiratory sensitiser, further data were requested by EFSA before reaching a final conclusion (2004).
The latest probiotic strain assessed was Bifidobacterium animalis (2012). For this strain (no further strain designation or details are available), the requirements regarding the assessment of antibiotic resistances were not met (as the strain was resistant to clindamycin and the genetic basis of the resistance could not be established). Based on two studies provided, the effect of B. animalis on GI-related parameters in dogs was considered of questionable biological relevance and EFSA could not conclude the efficacy of this product.
Apart from the strains mentioned above, other probiotics or synbiotics are available as nutritional supplements in dogs, both in Europe and in the USA. Even though most products available in Europe to date contain the E. faecium strain NCIMB 10415 E1707, sometimes in combination with other bacterial strains and different prebiotics, the products themselves have mostly not been specifically approved or tested. On the other hand, E. faecium NCIMB 10415 E1707 has been used most widely in experimental settings, to assess effect on immune function or gut health (see below). Other bacterial strains available as over-the-counter supplements for dogs contain different strains of Lactobacilli (L. acidophilus, L. casei, L. plantarum, L. paracasei, L. lactis, L. rhamnosus, L. salivarius), Bifidobacteria (B. infantis, B. lactis, B. longum, B. bifidum), Bacillus subtilis or coagulans and in some cases yeasts (Saccharomyces cerevisiae) or other fungi (Aspergillus oryzae). However, limited data are available about the safety and efficacy of these microorganisms/products or the health claims associated with them. Some microorganisms other than E. faecium have been tested as probiotics in experimental settings in dogs. For example Saccharomyces boulardii was investigated in a small pilot study presented as a congress abstract in dogs with IBD and protein-losing enteropathy (P-LE); (Bresciani et al. 2014)]. It significantly improved clinical activity score and serum albumin levels compared to the placebo-treated control dogs (Bresciani et al. 2014). Apart from single bacterial strains tested in vitro (detailed below), some single- and multi-strain probiotic products have also been tested to a certain degree in a clinical setting in dogs. For most single-strain studies, this is limited to the use of E. faecium (Swanson et al. 2002; Sauter et al. 2006; Strompfová et al. 2006; Schmitz et al. 2015a). Several probiotic cocktails have been used with variable effect. For example a mixture of lactobacilli that showed promising ex vivo results regarding creating a more tolerant microenvironment in the gut, did not significantly improve outcome when administered in a clinical trial (Sauter et al. 2006). In another study, strains from a product approved for the use in people (VSL#3) have been administered to dogs with IBD leading to some clinical and immunological improvement (Rossi et al. 2014). This includes four Lactobacilli (L. acidophilus, L. plantarum, L. paracasei, L. delbrueckii ssp. bulgaricus), three Bifidobacteria (B. breve, B. longum, B. infantis) and Streptococcus thermophilus.
Testing of microbial strains for their qualifications as probiotics in dogs
Several bacterial strains, mostly lactic acid producing bacteria (LAB) isolated from canine faeces, some originally used in other species or people, have been tested for their probiotic potential in vitro (but are currently not available in commercial products). Studies have especially focused on the survival properties of these strains at low pH (to mimic passage of the stomach), to resist degradation by bile acids in the small intestine, their adhesion properties to intestinal mucus and their potential to either produce antimicrobial peptides or to inhibit in vitro growth of pathogens (mostly E. coli and Salmonella ssp.). Some other functional and genetic properties (e.g. fermentation of carbohydrates, immune-modulating effects) have also been assessed. These studies and their main outcome are summarised in Table 1.
| Bacterial strain(s) | Source | Tested for | Result | Reference |
|---|---|---|---|---|
| Butyrivibrio fibrisolvens | Canine faeces | Presence and butyrate production | B. fibrisolvens was detected at low levels (2.4 × 103–9 × 105/g dry weight) in canine faeces and was only responsible for at most 30% of the butyrate production by mixed faecal microbes. Introduction of B. fibrisolvens at high number might increase butyrate and lactate production | Asanuma et al. (2001) |
| L. fermentum, L. mucosae, L. rhamnosus, L. salivarius, Weissella confusa | Canine faeces | MIC of 14 antibiotics, viability at pH2 for 4 h, antimicrobial activity against Micrococcus luteus | L. salivarius and W. confusa are possible probiotics | Beasley et al. (2006) |
| 16 isolates of Bifidobacterium animalis ssp. lactis | Canine faeces (GSD) | Survival in an in vitro digestion assay, viability at low pH, viability with bile salts, auto-aggregation activity | All strains showed good properties as potential probiotics | Bunešová et al. (2012) |
| L. fermentum, L. plantarum, L. rhamnosus and their combination | Unclear, but claimed to be ‘dog-specific’ | Adhesion to canine mucus, effect of different growth media (laboratory vs. manufacturing conditions), inactivation methods (80°C, 95°C and UV irradiation) | Better adhesion under laboratory conditions, inactivation by heat decreased adhesion properties | Grześkowiak et al. (2013) |
| Lactobacilli, Bifidobacteria and Enterococci | Canine large intestinal contents | Isolation frequency, similarity to known gene sequences | LAB are frequently found in canine faeces: 78% Lactobacillus, 11.6% Enterococcus, 6.8% Bifidobacterium, 2% Streptococcus bovis | Kim & Adachi (2007) |
| Enterococci | Dog faeces and dog feed | Adhesion to canine, porcine and human mucus | E. faecium and E. faecalis exhibit strain-dependent in vitro adhesion to human, canine and porcine mucus with no host specificity | Lauková et al. (2008) |
| Enterococci | 28 different commercially available dog feeds | Species identification, antibiotic sensitivity profiles, adhesion to human and canine mucus, lactic acid production, viability in bile | 22 selected strains classified: 6 E. faecium, 4 E. faecalis, 1 E. hirae, the remainder not classified. Good qualities as probiotics with no significant antibiotic resistance | Lauková et al. (2004) |
| Lactobacilli | Canine milk | Culture-based characterisation of lactobacilli (fermentation of carbohydrates, production of antimicrobial peptides, adhesion to mucin, MIC to antibiotics) | Some Lactobacillus sp. strains showed potential as probiotics | Martín et al. (2010) |
| Lactobacilli | Canine faeces | Bile resistance, inhibition of Salmonella typhimurium, production of reuterin | L. reuteri was dominant species, was more bile resistant, produced reuterin, inhibited Salmonella better than L. acidophilus | McCoy & Gilliland (2007) |
| Lactobacilli, Bifidobacteria | Canine intestinal mucosa-adherent microbiota (post-mortem) | pH sensitivity, bile resistance, pathogen inhibition, adherence to epithelial cells, survival after freeze-drying, feeding trial of B. animalis AHC7 | B. animals AHC7 showed best properties out of 62 strains of LAB isolated. It also reduced the carriage of Clostridium difficile in dogs | O'Mahony et al. (2009) |
| Lactobacilli | Canine faeces | pH sensitivity, bile resistance, inhibition of E.coli and Clostridium perfringens growth | Some isolates could colonise and persist in the GI tract and induce beneficial effects to the host | Perelmuter et al. (2008) |
| Lactobacilli | Canine faeces | In vivo administration of Lactobacillus murinus | L. murinus transiently persisted in the canine gut and is safe for administration | Perelmuter et al. (2011) |
| Lactobacillus acidophilus, Candida utilis enriched with selenium and zinc | Unknown | Blood selenium and zinc concentrations, blood antioxidant capacities, composition of intestinal microflora | Probiotic group: Increased blood selenium and zinc concentrations, increased activity of glutathione peroxidise, superoxide dismutase, total antioxidant capacity, increased Lactobacillus and Bifidobacterium count in the faeces | Ren et al. (2011) |
| L. rhamnosus GG, L. johnsonii, L. casei, L. bulgaris, L. pentosus | Bacterial strains: unknown, mucus: different species including dog | Adhesion of different probiotic LAB to different host mucus | Mucus adhesion properties are more dependent on the LAB strain than on the host | Rinkinen et al. (2003a) |
| LAB (L. rhamnosus GG, L. pentosus, B. lactis, E. faecium) | Unknown, some ‘canine’ origin | Inhibition of adhesion of canine and zoonotic pathogens (S. intermedius, S. typhimurium, C. perfringens, C. jejuni) to immobilised mucus from canine jejunal chyme | LAB of canine origin reduced adhesion of C. perfringens. No strain inhibited adhesion of S. typhimurium or S. intermedius. Both Enterococci enhanced adhesion of C. jejuni | Rinkinen et al. (2003b) |
| L. acidophilus (2 strains), L. johnsonii as a probiotic cocktail (PC) | Faecal samples from healthy dogs | PH-resistance, fermentation ability, anti-ETEC-activity, lactic acid/peroxide production, antibiotic resistance, storage capability, cytokine expression in canine PBMCs, cytokine expression in canine intestinal biopsies | PC increased IL-10 mRNA levels in healthy and inflamed tissues, especially in comparison to not altering TNFα, IFNγ and IL-12p40 levels → induction of a more tolerant micro-environment | Sauter et al. (2005) |
| Enterococcus faecium NCIMB 10415 E1707 | Commercial product | Cytokine production in canine intestinal biopsies and whole blood when co-cultured with EF or other TLR-ligands | Variety of changes in cytokine expression profile dependent on stimulant, TNFα increased in whole blood but reduced in biopsies, single TLR-ligands more potent and consistent than E. faecium stimulation | Schmitz et al. (2014) |
| Enterococcus faecium NCIMB 10415 E1707 | Commercial product | TNFα responses in whole blood compared to PBMCs from healthy dogs | E. faecium consistently produces TNFα responses, generally stronger response in whole blood than in PBMCs | Schmitz et al. (2013) |
| LAB | Faeces of 22 dogs | Characterisation of LAB (Bifidobacteria and Lactobacilli) in canine faeces (enzymatic activities, resistance to bile, antimicrobial susceptibility, inhibition of 15 indicator bacteria) | L. murinus, B. animalis and Pediococcus acidilactici most frequently isolated. All strains inhibited Gram-negative indicators (lactobacilli > bifidobacteria). L. reuteri showed best antimicrobial properties, resistance to bile observed in all strains, strain differences in pH-stability | Strompfová & Lauková (2014) |
| Lactobacilli, Enterococci | Faeces from 10 healthy dogs | Antimicrobial activity, tolerance to bile, adhesion properties | 40 strains of enterococci and 40 strains of lactobacilli isolated and tested, some showing potential as probiotics | Strompfová et al. (2004a) |
| Enterococci | Canine faeces | Bacteriocin production, pH and bile tolerance, antibiotic resistance, adhesion properties | Total count of 3.3–7.3 log10 CFU g−1 faeces, most strains were E. faecium, all sensitive to vancomycin, ampicillin, penicillin, chloramphenicol. 33% resistant to erythromycin, 28% to teteracycline. 75% showed broad inhibitor spectrum against Gram-positive indicator bacteria. Seven strains tested further: best probiotic properties: E. faecalis EE4 and E. faecium EF01 | Strompfová et al. (2004b) |
| Lactobacillus acidophilus | Canine jejunal chyme | Detection of specific L. acidophilus LAB20 strain using real-time PCR | LAB20 can be detected from canine faecal samples up to 6 weeks post-administration, could be candidate to study mechanism behind its persistence in the canine gut, could be probiotic candidate | Tang & Saris (2013) |
| Lactobacilli | Canine faeces | Effect of different carbon sources on the production of antimicrobial compounds against E.coli and S. typhimurium by 3 lactobacilli strains | Substrate affects the production of antimicrobial compounds by L. mucosae, L. acidophilus and L. reuteri in a dose- and pH- dependent manner | Tzortzis et al. (2004) |
| LAB | Canine faeces | Oxalate degradation by LABs, effect of different prebiotics (arabinogalactan, gum Arabic, lactitol, guar gum, inuin, maltodextrin, FOS) on oxalate degradation | 37 LAB were isolated, mean oxalate degradation was 17.7 ± 16.6%. The effect of prebiotics was variable, but overall greatest with guar gum. Manipulation of LAB might decrease intestinal oxalate, thus potentially reducing oxalate absorption and urolithiasis risk | Weese et al. (2004) |
Very little is known about the appropriate dose of probiotics in general in small animals, let alone in specific diseases. Survival characteristics of probiotic strains (especially E. faecium) have been tested in vitro and in vivo, that is in low environmental pH (mimicking gastric passage), the presence of bile, adhesion to mucus, recovery of live bacteria from faeces of dogs after oral administration (Lauková et al. 2004, 2008; Strompfová et al. 2004a; Marciňáková et al. 2006). Overall, it is not entirely clear if probiotic survival is even necessary for a beneficial effect, or if, for example their DNA is sufficient (Kant et al. 2014). There is some evidence that even non-viable probiotic bacteria can cause immune modulation in the host (Zhong et al. 2012). Also, there is very little information regarding possible interactions of strains in multi-strain formulations; even though some studies in experimental rodents or humans show a synergistic effect on the measured outcome (Baillon et al. 2004). The effect of formulation (liquid, capsule, in-feed) or natural sources of potential probiotics (yoghurt, raw green tripe, fermented plant material) is mostly unexplored to date. Some preliminary data on the effect of feeding raw meat, prebiotic fibre and yeast cell wall extract on the composition of faecal microbiota are available (Beloshapka et al. 2013). Changes depending on the meat protein source (chicken vs. beef) were less evident, but minor changes in faecal microbiota composition were seen when prebiotics were added (e.g. greater presence of fusobacteria, lower abundance of Faecalibacterium). The significance of these observed changes, however, remains unclear, especially as this study was performed in healthy dogs. Furthermore, most studies evaluated faecal samples and noticed only minor changes. However, recent studies suggest that probiotic mixtures (i.e. VSL#3) are able to induce major changes in the ileal mucosa-adherent microbiota in colitic mice and also dogs with chronic enteropathies (Mar et al. 2014; White et al. 2015).
Quality control is also an issue with probiotic products. As they are usually classified as nutritional supplements, quality control as for drugs is not legally required. Again, no large amount of data are available on the quality, shelf-life, etc., of commercially available probiotics. One study has assessed microbial components of dog food claiming to contain probiotics (Weese & Arroyo 2003). None of the 19 commercial diets contained all the claimed organisms, whereas one or more of the listed components could be isolated from around 50% of samples. Eleven samples contained additional, related organisms and more than 25% of tested diets showed no relevant bacterial growth. To the authors’ knowledge, no published study has so far assessed whether the claimed bacterial quality or quantity in probiotic nutritional supplements is according to label claims.
The effect of probiotics on selected parameters in healthy dogs
Enterococcus faecium
Interestingly, even though E. faecium is the most widely used probiotic strain in small animals, not many studies have focused on its safety or effects when administered to healthy dogs. In a study from 2003 performed at the Nestle Purina Product Technology Centre, puppies from different popular dog breeds were assigned one of two different diets after weaning (Benyacoub et al. 2003). One of the diets was a commercial, complete dog food with no supplements (control group), the other was the same diet with a stable encapsulated form of E. faecium 10415 SF68 in a dosage of 5 × 108 cfu day−1 added (treatment group). Main outcome measures included determination of total faecal IgA, total and vaccine-specific immunoglobulin gamma (IgG) and IgA serum concentrations, and quantification of circulating lymphocyte subsets by flow cytometry. Food intake, weight and routine laboratory parameters (complete blood count, serum biochemistry) were also evaluated. At the end of the study period (1 year), puppies consuming the test diet had significantly higher total faecal and serum IgA levels (but not serum IgG) compared to the control group. In addition, vaccination-associated IgA and IgG for canine distemper virus were also significantly higher in E. faecium-treated puppies compared to controls from week 31 on. All other parameters were not different between groups. This study concluded that E. faecium can enhance specific immune functions in young dogs (Benyacoub et al. 2003), but the clinical relevance is questionable as measured endpoints do not necessarily correlate to a ‘healthier’ intestine. Overall, evidence justifying the use of E. faecium under these conditions is limited.
In the second study investigating effects of E. faecium on healthy dogs, the strain used (E. faecium EE3) was isolated and enumerated from a commercially available canine food (Marciňáková et al. 2006). It was orally administered to healthy adult dogs of different breeds and ages for 1 week at a daily dosage of 2–3 × 109 cfu dog−1. Faecal cultures and blood samples for routine biochemistry values were obtained before and at the end of the treatment period, as well as 1, 2 and 3 months after cessation of probiotic administration. E. faecium treatment did not cause any clinical side-effects. The strain persisted in faeces for 3 months after cessation of treatment (reaching average concentrations of 6.83 ± 0.95 log cfu g−1). Total concentration of LAB increased, and Pseudomonas-like bacteria and Staphylococcus spp. decreased in faecal samples, but the abundance of E. coli was not influenced. Total serum lipids and protein decreased in most dogs with treatment, with cholesterol being within the reference range of all dogs at the end of the treatment period. This study, therefore, inferred a beneficial influence of E. faecium on canine health, possibly even in obesity, even though obese animals were not examined in the study, and the beneficial effect of normalisation of cholesterol is questionable (Marciňáková et al. 2006). In addition, this is the only study that has shown long-term persistence of orally administered probiotics in dogs, there was a lack of a control group and it is not entirely clear how the strain identification was performed; hence the results of this study have to be interpreted with caution. Also, for both of these studies, it has to be questioned if the definition of probiotics has been fulfilled using E. faecium, as a relevant benefit of increased faecal IgA, elevated vaccine-associated titres or ‘lowered’ cholesterol has not been demonstrated.
Lactobacilli and bifidobacteria
A number of studies have investigated the safety and effect of single-strai n LAB preparations in healthy dogs. For example effects on measured outcomes of the administration of L. acidophilus have been controversial. In one study, its administration (L. acidophilus DSM13241) to adult dogs was associated with changes in haematological and immunological parameters (increased red blood cells [RBC], Hct, haemoglobin, neutrophils, monocytes and serum IgG, reduced RBC fragility and serum NO concentrations) of questionable clinical relevance (Baillon et al. 2004). In another study, the supposedly beneficial changes observed in the composition of the microbiota and faecal metabolites was more attributed to the addition of prebiotic FOS than to L. acidophilus NCFM itself (Swanson et al. 2002).
L. fermentum has been tested by the same group in two studies (Strompfová et al. 2006, 2012a). This LAB (L. fermentum AD1) was originally isolated from faeces of a healthy dog (6-year-old Tibetan Terrier) and was shown to have good in vitro survival at a pH of 3.0 for 3 h (86.8%), and in the presence of 1% bile (75.4%), as well as good adhesion properties to canine and human intestinal mucus, and no unacceptable antimicrobial resistance (Strompfová et al. 2006). It was orally administered to 15 healthy dogs of various breeds at a dose of 3 × 109 cfu dog−1 for 7 days. Significant increases of faecal lactobacilli, enterococci, and serum total protein, total lipids and reduction in blood glucose were noted. In a follow-up study, the strain was administered in freeze-dried form to healthy dogs, and was shown to persist short-term in the GI tract and to increase SCFA concentrations. Additionally, a reduction in Clostridia and Gram-negative bacteria (coliforms, Aeromonas, Pseudomonas) were also noted by faecal culture (Strompfová et al. 2012a). In the study, this was implied as a desired outcome, however, as this is a culture-based approach and some members of Clostridia have been identified as part of the normal beneficial gut flora (see above), deductions regarding gut health are difficult to make from these data. More detailed investigations to evaluate whether this probiotic strain reduces specifically the potential pathogen C. perfringens rather than Clostridia in general would be needed.
Lactobacillus animalis LA4 (isolated from the faeces of a healthy adult dog) was tested in vitro and in vivo in nine dogs (freeze-dried, given for 10 days). On day 11, cultured faecal lactobacilli concentrations were increased and enterococci reduced compared to the beginning of the trial, hence this strain was concluded to have some probiotic properties (Biagi et al. 2007). However, this conclusion is nearly impossible to make, as semi-quantitative culture was the only outcome assessed and there was no control group.
A more recent study used a genetically engineered strain of Lactobacillus casei (no further strain designation available), capable of producing biologically active canine granulocyte macrophage colony stimulating factor, and investigated its properties as a probiotic for dogs (Chung et al. 2009). It was administered at 1 × 109 cfu day−1 for 7 weeks. Treated dogs showed increased monocyte counts, serum IgA and canine coronavirus-specific vaccination-associated IgG compared with dogs fed a regular diet without probiotic supplements and dogs receiving a non-engineered strain of L. casei (Chung et al. 2009). The clinical relevance of this finding and the safety and efficacy of administration of this strain remains open.
Different strains of Bifidobacterium animalis [AHC7; (Kelley et al. 2010); and an unspecified strain isolated from dog faeces (Strompfová & Lauková 2014)], were investigated by the same group and found to not cause any undesirable effects. Their administration to healthy dogs increased the number of cultured LAB, but lowered the count of coliform bacteria in canine faeces. Other faecal and serum parameters as well as the phagocytic activity of peripheral blood leucocytes (especially neutrophils) were improved in treated dogs compared with the untreated control group. These effects could be detected several weeks after the treatment had been ceased (Strompfová et al. 2014), but again their relevance remains unclear, especially as methods used were rather crude assessments of immune function.
Overall data on improving gut health or immunological status in dogs using lactobacilli or bifidobacteria are not compelling, especially in the light of the fact that it is not known if increasing certain bacterial phyla is correlated with improved GI function or a lower incidence of diarrhoeic diseases.
Bacilli
Bacilli are thought by some authors to represent superior probiotics to LABs, as they can sporulate and thus be more resistant to environmental stress and low pH (Biourge et al. 1998; Félix et al. 2010). However, mere survival might be not the most important feature of a probiotic, and they all need to be tested for their benefit in clinical situations. In some European countries, probiotic products containing Bacilli are available as nutritional supplements for humans and animals (Biourge et al. 1998). They have been shown to have beneficial effects on the survival of mice infected with Klebsiella pneumoniae and on the breeding performances of a number of production animals (Biourge et al. 1998). Bacillus CIP 5832 was found to be persistent when added to canine food and when exposed to expansion-extrusion and drying experiments. They were also able to survive the canine GI tract, however, did not seem to persist, as they disappeared from faeces 3 days after cessation of administration (German et al. 2000). Only one study found that Bacillus subtilis C-3102 can improve faecal texture and odour in dogs due to a decreased content of faecal ammonia (Félix et al. 2010). Once again, the clinical relevance of this finding and the justification of calling bacilli ‘probiotics’ in this situation is highly questionable and the usage of Bacilli as probiotics cannot be recommended.
Probiotic mixtures
Mixtures of probiotics used in healthy dogs have been of variable composition; in addition, outcome measures were different, mainly also due to the availability of different techniques to assess changes in microbial communities. One study administered five potentially probiotic LAB strains (L. fermentum, L. salivarius, Weissella confuse, L. rhamnosus and L. mucosae) to five permanently fistulated Beagle dogs for 7 days (Manninen et al. 2006). Denaturing gradient gel electrophoresis (DGGE) demonstrated that the LAB modified the dominant indigenous jejunal LAB microbiota. All strains were undetectable 7 days after administration ceased and effects were transient. In another study (Garcia-Mazcorro et al. 2011), seven strains of LAB (E. faecium, S. salivarus ssp. thermophilus, B. longum, L. acidophilus, L. casei ssp. rhamnosus, L. plantarum, L. delbrueckii ssp. bulgaricus) were administered to 12 healthy dogs and faecal microbial communities were assessed using DGGE gels, 16S rRNA gene libraries, quantitative PCR and 16S rRNA gene 454-pyrosequencing. Probiotic species were detectable in 11/12 dogs during product administration, but not before or after. Abundances of Enterococcus and Streptococcus spp. were significantly increased. However, on pyrosequencing, no changes in the major bacterial phyla were observed. This study concluded that the product was well tolerated and did not cause any clinical side-effects. Administration of the product resulted in increased abundance of the probiotic genera, but this was not sufficient to cause significant changes in the overall microbiome structure (Garcia-Mazcorro et al. 2011) and certainly cannot automatically be inferred to convey a health benefit.
Finally, a synbiotic consisting of E. faecium SF68, Bacillus coagulans, L. acidophilus and several prebiotics (FOS, MOS) and vitamins (B3, B6) was administered in a placebo-controlled trial to healthy trained sled dogs, and changes of the composition of the faecal microbiota assessed using quantitative PCR and tag-encoded FLX 16S rDNA amplicon pyrosequencing (Gagné et al. 2013). Alterations in the faecal microbiome observed included a rise in Lactobacillaceae and an increased faecal butyrate concentration across all dogs. Faecal scores also improved compared with the control group at 5 weeks (Gagné et al. 2013). Whether these findings are correlated, beneficial to the host or even only incidental, remains unclear.
Use of probiotics in small animal gastrointestinal diseases
Infectious and non-infectious acute diarrhoea
Overall, it seems that – possibly depending on the probiotic strain or mixture used – there is some merit in the use of probiotics in acute infectious canine GI diseases. Administration of the probiotic mixture VSL#3 in a randomised manner to puppies with confirmed parvoviral enteritis lead to an increased percentage of surviving dogs (90% in probiotic group vs. 70% in the non-probiotic group), and a more rapid improvement of clinical scores and leucocyte/lymphocyte counts (Arslan et al. 2012). In another study, there was a significant reduction in Ancylostoma eggs shedding in 10 dogs treated with a mixture of LABs (L. acidophilus ATCC 4536, L. plantarum ATCC 8014 and L. delbrueckii UFV H2B20) for 28 days compared with an untreated control group (Coêlho et al. 2013). Similar results could not be achieved for the treatment of canine giardiasis with E. faecium SF68: after 6 weeks of treatment no differences in cyst shedding, faecal antigen shedding, faecal IgA or leucocyte phagocytic activity were observed between treated and untreated dogs (Simpson et al. 2009).
Other forms of acute diarrhoea in dogs in which probiotics have been administered include stress-associated (e.g. kennelling stress), antibiotic-induced and idiopathic diarrhoea. Results are variable, depending on the probiotic strain and the dog population evaluated. Using E. faecium SF68, there was no effect on kennel stress-associated diarrhoea, which might partially be due to the low prevalence of diarrhoea in this study (Bybee et al. 2011). Faecal scores were significantly improved in dogs undergoing kennelling stress when supplemented with Bifidobacterium animalis AHC7 compared with an untreated control group (Kelley et al. 2012). The same strain was able to significantly reduce the time to resolution of clinical signs and the number of dogs receiving metronidazole in a study of acute idiopathic diarrhoea (dose of 2 × 1010 cfu day−1) (Kelley et al. 2009). Similar responses were seen in a study of acute gastroenteritis using a probiotic mixture of L. acidophilus, Pediococcus acidilactici, B. subtilis, B. licheniformis and L. farciminis (Herstad et al. 2010). Recovery time was significantly reduced (mean 1.3 days, 95% CI: 0.5–2.1 days) compared with untreated controls (mean 2.2 days, 95% CI: 1.3–3.1 days) with a comparably large dose of 4.2 × 109 cfu/10 kg three times daily (Herstad et al. 2010). Inactivated bacterial compounds as part of an ‘enterovaccine’ might also be useful in treating recurrent self-limiting episodes of diarrhoea (e.g. stress-related) in dogs. One commercially available preparation (deactivated whole bacteria and lysates of E.coli, Bacillus pumilus, Morganella morganii, Alcaligenes faecalis, Shigella flexneri, Bacillus subtilis, Enterococcus faecalis and Proteus vulgaris) reduced the number of diarrhoea episodes and severity in five of six treated dogs (Cerquetella et al. 2012). There were no control dogs in this pilot study, hence the exact potential of this inactivated bacterial mixture needs to be evaluated by further clinical studies. Interestingly, administration of Saccharomyces boulardii to dogs with experimental lincomycin-induced diarrhoea could prevent, but not treat this condition (Aktas et al. 2007).
Chronic diarrhoea
Similar to people, the pathogenesis of chronic inflammatory conditions of the GI tract in dogs (e.g. CE/IBD) is assumed to be due to an aberrant response of the immune-system to the luminal or adherent intestinal microbiota (Sartor 2006; Hall & German 2010). There is ample evidence of immune dysregulation, even though the exact type of inflammatory response and pathogenesis has not been elucidated yet (German et al. 2000; Peters et al. 2005; Jergens et al. 2009; Schmitz et al. 2012). Several studies have shown that there are also alterations of the intestinal microbiome present in canine CE/IBD (Suchodolski et al. 2012a, b). Hence, there have been several attempts to influence the composition of the microbiota in those dogs to alleviate clinical signs; partially using probiotics that had already shown to have some immune-modulatory properties in in vitro or ex vivo studies (Sauter et al. 2005, 2006; Schmitz et al. 2013, 2014). E. faecium NCIMB 10415 E1707 has been assessed as a single-strain treatment in dogs with food-responsive disease (FRD) and found to have no effect on clinical activity score, histology scores or duodenal and colonic gene expression of selected genes associated with specific T-helper lymphocyte lines (Schmitz et al. 2015b). In addition, there was also no effect of E. faecium treatment on gene or protein expression of inflammasome compounds (Schmitz et al. 2015b). More promising results could be achieved by using probiotic mixtures in FRD dogs: A combination of LAB (L. acidophilus and L. johnsonii) reduced duodenal interleukin (IL)-10 and colonic interferon gamma (IFNγ) mRNA levels and the number of faecal Enterobacteriaceae, whereas numbers of Lactobacillus spp. increased. Clinical improvement was noted to similar levels in dogs receiving the LAB cocktail compared with dogs treated with diet alone (Sauter et al. 2005).
Additionally, a probiotic mixture with VSL#3 strains formulated for pets (SIVOY™; details above) was used in dogs with idiopathic IBD and compared to a treatment regimen with metronidazole and prednisolone in an open-label trial (Rossi et al. 2014). Clinical activity, duodenal histology scores and CD3+ lymphocytes in the intestinal tissue decreased post-treatment in both groups. However, FoxP3+ cells (a marker of regulatory T-helper lymphocytes [Tregs]) increased significantly after treatment only in the dogs treated with VSL#3 strains. Also, transforming growth factor beta (TGFb)+ cells (most likely Tregs) increased in both groups after treatment, but to a greater magnitude in probiotic treated dogs. There was some effect of the probiotics on expression of tight junction proteins, with occludin being significantly elevated in healthy control dogs and dogs treated with probiotics compared with IBD dogs. Microbiome analysis based on quantitative PCR revealed a reduced abundance of Faecalibacterium and Turicibacter in dogs with IBD at the start of the trial, with a significant increase in Faecalibacterium observed in the animals treated with VSL#3 strains (Rossi et al. 2014). This is noteworthy, as Faecalibacterium prausnitzii has been advocated as an anti-inflammatory commensal bacterium in people (Miquel et al. 2013) and is of lower abundance in intestinal contents or faecal samples from human and canine IBD patients (Suchodolski et al. 2012b; Fujimoto et al. 2013).
Saccharomyces boulardii yeasts have been administered to dogs with CE/PLE and healthy control dogs in the small pilot placebo-controlled double-blinded clinical trial mentioned above at 1 × 109 cfu kg−1 body weight BID for 10 days (Bresciani et al. 2014). Dogs with CE or PLE additionally received standard medical treatment consisting of diet, antibiotics and/or immunosuppressive drugs. S. boulardii was not detected in faecal samples of healthy dogs before treatment started, but was present after 1 day of supplementation, reached highest levels after 5 days (10 × 107 cfu g−1) and was eliminated 4 days after withdrawal of treatment. In dogs with CE, clinical score improved significantly, and in dogs with PLE serum albumin values increased significantly compared with placebo treatment. Duodenal endoscopic and histology scores were not different before and after treatment in any of the dogs. The study concluded that S. boulardii can be safely administered to dogs and it might be useful as an adjunctive treatment in CE and PLE (Bresciani et al. 2014). However, as it is a small study and has not been fully published yet, awaiting of the full results and further research into the usefulness of S. boulardii as a potential probiotic is warranted.
Faecal microbial transplants
Administration of single-strain or even multi-strain probiotic products might have a limited ability to influence the composition of the intestinal microbiome permanently, especially given the fact that the microbiome consists of hundreds of microbial species. Based on the assumption that a more complex change is needed in certain conditions (e.g. Clostridium difficile infection or IBD), there have been attempts of transferring the intestinal microbiota from one subject to another. This has been termed faecal microbial transplantation (FMT), microbiome restorative therapy or faecal bacteriotherapy. In human medicine, FMT has been mostly performed for recurrent C. difficile infection. Two reviews show that it is safe and effective in 83–92% of human patients, which achieved full resolution of clinical signs (Gough et al. 2011; Guo et al. 2012). Limited clinical data are available evaluating the use of FMT for IBD in humans, but pilot studies suggest that the response rate to FMT in chronic intestinal inflammation is much lower compared with C. difficile infection (Colman & Rubin 2014). Experience in small animals regarding the safety and efficacy of FMT is scarce and anecdotal. Two congress abstracts report some preliminary findings in dogs. One is a case report of an ongoing study (not published), where a dog with eosinophilic IBD of 2 years duration and moderate clinical signs with conventional therapy was given the FMT by enema, with 45 min retention time. Faecal consistency improved within 24 h and the dog had been clinically well at the time of the report (3 months after treatment). Next-generation 16S rRNA gene sequencing of the faecal microbiome revealed that by day 2 after FMT, the dogs’ faecal sample clustered with the donor, not the own baseline sample, and species richness increased compared to pre-treatment values (Weese 2013). The other abstract reports the use of FMT in 8 dogs with refractory presumptive Clostridium perfringens-associated diarrhoea (Murphy et al. 2014). Again, it was administered as an enema (1–3 transplants per dog). All dogs had immediate resolution of their diarrhoea and 6/8 dogs were negative on follow-up PCR panels for C. perfringens alpha toxin. As mentioned before, C. perfringens can be part of the normal intestinal flora in dogs, hence it remains unclear if the detected C. perfringens was really the cause of the diarrhoea or rather a part of intestinal dysbiosis (Minamoto et al. 2014b). If that had been the case, it needs still to be critically assessed whether the FMT addressed Clostridiosis, even if the microbiome changes suggest an improvement of faecal dysbiosis. Both authors of the unpublished conference abstracts conclude that FMT should be considered as a treatment option in dogs failing other therapeutic options (Weese 2013; Murphy et al. 2014) and anecdotal evidence of its usefulness is increasing in the veterinary community. Prospective studies in large cohorts of dogs are needed to properly address the usefulness of FMT in defined GI diseases in dogs.
Conclusions
The microbiota on mucosal surfaces, especially the GI tract, in dogs is complex. The composition varies from site to site and there is some evidence that changes in the composition of the microbiota/microbiome are associated with certain diseases. However, analysis of the canine GI and faecal microbiota composition, its function, production of metabolites and immunological properties is far from complete, even though data on the microbiome are accumulating. Accordingly, overall knowledge of the best characteristics of a canine microbial commensal or probiotic is patchy and most assumptions about their best properties are derived from human studies. There are some bacterial strains that show promise as potential probiotics, especially LAB. However, the effects of the most commonly used strain (E. faecium) are still not well understood, particularly in diseased dogs. It is challenging to compare outcomes of different studies, both in healthy and in diseased dogs, as there is huge variation in the probiotics used (single-strain, multi-strain, type of microorganism), their dosage, application form and frequency. Measured outcomes are also not consistent. Overall, it seems to the authors that E. faecium tends to be more suitable for acute and/or infectious forms of diarrhoea; as there is some evidence it produces a more pro-inflammatory rather than anti-inflammatory reaction (Schmitz et al. 2014, 2015b); whereas some lactobacilli and bifidobacteria show more pronounced immune-regulatory functions. Intriguingly, especially a combination of LABs (VSL#3) used in human medicine to prevent relapse of Ulcerative Colitis has some promise in treating chronic enteropathies in dogs and in creating a more anti-inflammatory local environment (Rossi et al. 2014). It is interesting to note that even in well-defined infectious diseases like parvovirosis or parasitic infestation, some probiotics show a beneficial effect. The mechanism behind this is not well understood and further research is warranted. It is clear from studies in people and experimental rodents, that the immunological outcome depends on both the bacterial strain or even subspecies – probably in a dose-dependent manner (Weese & Anderson 2002; Evrard et al. 2011; Garcia-Mazcorro et al. 2011) – and the respective hosts immune response. In humans, probiotics have been classified into ‘pro-inflammatory’ and ‘anti-inflammatory’ by some authors, depending on their major properties (Shida et al. 2011). Because of this, careful assessment of potential probiotics in the target species and disease – possibly both in vitro and in vivo – is necessary to judge their full potential. Some authors even propose to test all probiotics in vitro or in tissue explants first, before performing in vivo trials (Tsilingiri et al. 2012). However, this might not always be possible in veterinary medicine.
There should also be a consideration that bacteria might not always be the most appropriate probiotics. We know virtually nothing regarding the fungal or even viral composition of the normal intestinal tract or other mucosal surfaces in dogs (Foster et al. 2013). There is some evidence that yeasts like Saccharomyces boulardii might be valid alternative probiotics and need more detailed investigations. Furthermore, an emerging field is the research into bacterial metabolites (e.g. indole, acetate) that may potentially serve as postbiotics.
Faecal transplantation is an interesting option to treat both acute and chronic diarrhoea in dogs, however, much more needs to be understood regarding its best performance, safety and usefulness, before it can be recommended as a routine treatment.
In summary, more work needs to be done to understand the complex interplay between potential probiotics and their host environment. Very careful investigations into mechanisms of action and detailed measured outcomes will help to understand which probiotic is useful in which condition, and which changes of the intestinal microbiome are necessary to achieve clinical remission of both acute and chronic conditions.
Acknowledgments
This manuscript was not supported by any funding body.
Source of funding
This manuscript has not been supported by any funding.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Contributions
Both authors contributed equally to this manuscript.
References
- Aktas M.S., Borku M.K. & Ozkanlar Y. (2007) Efficacy of Saccharomyces boulardii as a probiotic in dogs with lincomycin induced diarrhoea. Bulletin of the Veterinary Institute in Pulawy51, 365–369.
- Allaart J.G., van Asten A.J.A.M., Vernooij J.C.M. & Gröne A. (2011) Effect of Lactobacillus fermentum on beta2 toxin production by Clostridium perfringens. Applied and Environment Microbiology77, 4406–4411.
- Arslan H.H., Aksu D.S., Terzi G. & Nisbet C. (2012) Therapeutic effects of probiotic bacteria in parvoviral enteritis in dogs. Revue De Medecine Veterinaire163, 55–59.
- Asanuma N., Kawato M. & Hino T. (2001) Presence of Butyrivibrio fibrisolvens in the digestive tract of dogs and cats, and its contribution to butyrate production. Journal of General and Applied Microbiology47, 313–319.
- Baillon M.L., Marshall-Jones Z.V. & Butterwick R.F. (2004) Effects of probiotic Lactobacillus acidophilus strain DSM13241 in healthy adult dogs. American Journal of Veterinary Research65, 338–343.
- Bayoumi M.A. & Griffiths M.W. (2012) In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. International Journal of Food Microbiology156, 255–263.
- Beasley S.S., Manninen T.J.K. & Saris P.E.J. (2006) Lactic acid bacteria isolated from canine faeces. Journal of Applied Microbiology101, 131–138.
- Bell J.A., Kopper J.J., Turnbull J.A., Barbu N.I., Murphy A.J. & Mansfield L.S. (2008) Ecological characterization of the colonic microbiota of normal and diarrheic dogs. Interdisciplinary Perspectives on Infectious Diseases2008, 149694.
- Beloshapka A.N., Dowd S.E., Suchodolski J.S., Steiner J.M., Duclos L. & Swanson K.S. (2013) Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiology Ecology84, 532–541.
- Benyacoub J., Czarnecki-Maulden G.L., Cavadini C., Sauthier T., Anderson R.E., Schiffrin E.J.et al. (2003) Supplementation of food with Enterococcus faecium (SF68) stimulates immune functions in young dogs. Journal of Nutrition133, 1158–1162.
- Biagi G., Cipollini I., Pompei A., Zaghini G. & Matteuzzi D. (2007) Effect of a Lactobacillus animalis strain on composition and metabolism of the intestinal microflora in adult dogs. Veterinary Microbiology124, 160–165.
- Biourge V., Vallet C., Levesque A., Sergheraert R., Chevalier S. & Roberton J.L. (1998) The use of probiotics in the diet of dogs. Journal of Nutrition128 (12 Suppl.), 2730S–2732S.
- Bresciani F., D'Angelo S., Fracassi F., Galuppi R., Diana A., Linta N.et al. (2014) Efficacy of Saccharomyces boulardii in the treatment of dogs with chronic enteropathies - randomized double-blind placebo-controlled study. European College of Veterinary Internal Medicine - Companion Animals Congress Proceedings. on www.vin.com.
- Bunešová V., Vlková E., Rada V., Ročková Š., Svobodová I., Jebavý L.et al. (2012) Bifidobacterium animalis subsp. lactis strains isolated from dog faeces. Veterinary Microbiology160, 501–505.
- Busch K., Suchodolski J.S., Kühner K.A., Minamoto Y., Steiner J.M., Mueller R.S.et al. (2015) Clostridium perfringens enterotoxin and Clostridium difficile toxin A/B do not play a role in acute haemorrhagic diarrhoea syndrome in dogs. The Veterinary Record176, 253.
- Bybee S.N., Scorza A.V. & Lappin M.R. (2011) Effect of the probiotic Enterococcus faecium SF68 on presence of diarrhea in cats and dogs housed in an animal shelter. Journal of Veterinary Internal Medicine25, 856–860.
- Callaway T.R., Edrington T.S., Anderson R.C., Harvey R.B., Genovese K.J., Kennedy C.N.et al. (2008) Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Animal Health Research Reviews9, 217–225.
- Castagliuolo I., Riegler M.F., Valenick L., LaMont J.T. & Pothoulakis C. (1999) Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infection and Immunity67, 302–307.
- Cerquetella M., Laus F., Speranzini F., Carnevali C., Spaterna A., Battaglia E.et al. (2012) Efficacy of an enterovaccine in recurrent episodes of diarrhea in the dog: a pilot study. Revista Espanola de Enfermedades104, 65–68.
- Chung J.Y., Sung E.J., Cho C.G., Seo K.W., Lee J.S., Bhang D.H.et al. (2009) Effect of recombinant Lactobacillus expressing canine GM-CSF on immune function in dogs. Journal of Microbiology and Biotechnology19, 1401–1407.
- Coêlho M.D.G., Coêlho F.A.D.S., de Mancilha I.M., (2013). Probiotic therapy: a promising strategy for the control of canine hookworm. Journal of Parasitology Research2013. doi: 10.1155/2013/430413.
- Collado M.C., Grześkowiak Ł. & Salminen S. (2007a) Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Current Microbiology55, 260–265.
- Collado M.C., Meriluoto J. & Salminen S. (2007b) Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Letters in Applied Microbiology45, 454–460.
- Colman R.J. & Rubin D.T. (2014) Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. Journal of Crohns and Colitis8, 1569–1581.
- Culligan E.P., Hill C. & Sleator R.D. (2009) Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathogens1, 19.
- Cummings JH. Short chain fatty acids in the human colon. Gut1981; 22: 763–779.
- Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. Journal of Applied Bacteriology1991; 70: 443–459.
- Davis CP, Cleven D, Balish E, Yale CE. Bacterial association in the gastrointestinal tract of beagle dogs. Applied and Environment Microbiology1977; 34: 194–206.
- Evrard B, Coudeyras S, Dosgilbert A, Charbonnel N, Alamé J, Tridon A, et al. Dose-dependent immunomodulation of human dendritic cells by the probiotic Lactobacillus rhamnosus Lcr35. PLoS ONE2011; 6: e18735.
- FAO/WHO (2002) Working group for drafting guidelines for the evaluation of probiotics in food. Available at : ftp://ftpfaoorg/es/esn/food/wgreport2pdf 2002.
- Félix AP, Netto MVT, Murakami FY, De Brito CBM, De Oliveira SG, Maiorka A. Digestibility and fecal characteristics of dogs fed with Bacillus subtilis in diet. Ciência Rural2010; 40: 2169–2173.
- Fijan S. (2014) Microorganisms with claimed probiotic properties: an overview of recent literature. International Journal of Environmental Research and Public Health11, 4745–4767.
- Foster M., Dowd S.E., Stephenson C.E., Steiner J.M., Suchodolski J.S. (2013). Characterization of the fungal microbiome (mycobiome) in fecal samples from dogs. Journal of Veterinary Internal Medicine doi: 10.1155/2013/658373.
- Fujimoto T., Imaeda H., Takahashi K., Kasumi E., Bamba S., Fujiyama Y.et al. (2013) Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohns disease. Journal of Gastroenterology and Hepatology28, 613–619.
- Gagné J.W., Wakshlag J.J., Simpson K.W., Dowd S.E., Latchman S., Brown D.A.et al. (2013) Effects of a synbiotic on fecal quality, short-chain fatty acid concentrations, and the microbiome of healthy sled dogs. BMC Veterinary Research9, 246.
- Garcia-Mazcorro J.F., Lanerie D.J., Dowd S.E., Paddock C.G., Grützner N., Steiner J.M.et al. (2011) Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiology Ecology78, 542–554.
- Garcia-Mazcorro J.F., Dowd S.E., Poulsen J., Steiner J.M. & Suchodolski J.S. (2012) Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiologyopen1, 340–347.
- German A.J., Helps C.R., Hall E.J. & Day M.J. (2000) Cytokine mRNA expression in mucosal biopsies from German shepherd dogs with small intestinal enteropathies. Digestive Diseases and Sciences45, 7–17.
- Ghouri Y.A., Richards D.M., Rahimi E.F., Krill J.T., Jelinek K.A. & DuPont A.W. (2014) Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clinical and Experimental Gastroenterology7, 473–487.
- Gibson G.R. & Roberfroid M.B. (1995) Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. Journal of Nutrition125, 1401–1412.
- Gibson G.R., Scott K.P., Rastall R.A., Tuohy K.M., Hotchkiss A., Dubert-Ferrandon A.et al. (2010) Dietary prebiotics: current status and new definition. Food Science and Technology Bulletin7, 1–19.
- Gough E., Shaikh H. & Manges A.R. (2011) Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clinical Infectious Diseases53, 994–1002.
- Grześkowiak L., Endo A., Collado M.C., Pelliniemi L.J., Beasley S. & Salminen S. (2013) The effect of growth media and physical treatments on the adhesion properties of canine probiotics. Journal of Applied Microbiology115, 539–545.
- Guard B.C., Barr J.W., Reddivari L., Klemashevich C., Jayaraman A., Steiner J.M.et al. (2015) Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS ONE10, e0127259.
- Guo B., Harstall C., Louie T., Veldhuyzen van Zanten S. & Dieleman L.A. (2012) Systematic review: faecal transplantation for the treatment of Clostridium difficile-associated disease. Alimentary Pharmacology & Therapeutics35, 865–875.
- Hall E.J. (2011) Antibiotic-responsive diarrhea in small animals. The Veterinary Clinics of North America: Small Animal Practice41, 273–286.
- Hall E.J., German A.J. (2010) Diseases of the small intestine. In: Texbook of Veterinary Internal Medicine. (eds S.J. Ettinger & E.C. Feldman), 7th edn, pp 1560–1565. Expert Consult: St. Louis, Missouri, USA.
- Handl S., Dowd S.E., Garcia-Mazcorro J.F., Steiner J.M. & Suchodolski J.S. (2011) Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiology Ecology76, 301–310.
- Handl S., German A.J., Holden S.L., Dowd S.E., Steiner J.M ., Heilmann R.M.et al. (2013) Faecal microbiota in lean and obese dogs. FEMS Microbiology Ecology84, 332–343.
- Hang I., Rinttila T., Zentek J., Kettunen A., Alaja S., Apajalahti J.et al. (2012) Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Veterinary Research8, 90.
- Herstad H.K., Nesheim B.B., L'Abée-Lund T., Larsen S. & Skancke E. (2010) Effects of a probiotic intervention in acute canine gastroenteritis - A controlled clinical trial. Journal of Small Animal Practice51, 34–38.
- Honneffer J.B., Minamoto Y. & Suchodolski J.S. (2014) Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World Journal of Gastroenterology20, 16489–16497.
- Hooda S., Minamoto Y., Suchodolski J.S. & Swanson K.S. (2012) Current state of knowledge: the canine gastrointestinal microbiome. Animal Health Research Reviews13, 78–88.
- Hughes R. & Rowland I.R. (2001) Stimulation of apoptosis by two prebiotic chicory fructans in the rat colon. Carcinogenesis22, 43–47.
- Hutchins R.G., Bailey C.S., Jacob M.E., Harris T.L., Wood M.W., Saker K.E.et al. (2013) The effect of an oral probiotic containing lactobacillus, bifidobacterium, and bacillus species on the vaginal microbiota of spayed female dogs. Journal of Veterinary Internal Medicine27, 1368–1371.
- Jergens A.E., Sonea I.M., O'Connor A.M., Kauffman L.K., Grozdanic S.D., Ackermann M.R.et al. (2009) Intestinal cytokine mRNA expression in canine inflammatory bowel disease: a meta-analysis with critical appraisal. Comparative Medicine59, 153–162.
- Jia J., Frantz N., Khoo C., Gibson G.R., Rastall R.A. & McCartney A.L. (2010) Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiology Ecology71, 304–312.
- Jones S.E. & Versalovic J. (2009) Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiology9, 35.
- Kant R., de Vos W.M., Palva A. & Satokari R. (2014) Immunostimulatory CpG motifs in the genomes of gut bacteria and their role in human health and disease. Journal of Medical Microbiology63 (Pt 2), 293–308.
- Kelley R.L., Minikhiem D., Kiely B., O'Mahony L., O'Sullivan D., Boileau T.et al. (2009) Clinical benefits of probiotic canine-derived Bifidobacterium animalis strain AHC7 in dogs with acute idiopathic diarrhea. Veterinary Therapeutics : Research in Applied Veterinary Medicine10, 121–130.
- Kelley R., Soon Park J., O'Mahony L., Minikhiem D. & Fix A. (2010) Safety and tolerance of dietary supplementation with a canine-derived probiotic (bifidobacterium animalis strain AHC7) fed to growing dogs. Veterinary Therapeutics : Research in Applied Veterinary Medicine11, E1–E14.
- Kelley R., Levy K., Mundell P. & Hayek M.G. (2012) Effects of varying doses of a probiotic supplement fed to healthy dogs undergoing kenneling stress. International Journal of Applied Research in Veterinary Medicine10, 205–216.
- Kim S.-Y. & Adachi Y. (2007) Biological and genetic classification of canine intestinal lactic acid bacteria and bifidobacteria. Microbiology and Immunology51, 919–928.
- Koh J.H., Choi S.H., Park S.W., Choi N.J., Kim Y. & Kim S.H. (2013) Synbiotic impact of tagatose on viability of Lactobacillus rhamnosus strain GG mediated by the phosphotransferase system (PTS). Food Microbiology Elsevier Ltd; 36, 7–13.
- Lauková A., Strompfová V. & Ouwehand A. (2004) Adhesion properties of enterococci to intestinal mucus of different hosts. Veterinary Research Communications28, 647–655.
- Lauková A., Marciĕáková M., Strompfová V. & Ouwehand A.C. (2008) Probiotic potential of enterococci isolated from canine feed. Folia Microbiologica53, 84–88.
- Lederberg J., McCray A. (2001) ‘Ome Sweet ‘Omics - A Genealogical Treasury of Words. Scientist Inc.: Philadelphia, PA, 15, 8.
- Lee Y.-K., Puong K.-Y., Ouwehand A.C. & Salminen S. (2003) Displacement of bacterial pathogens from mucus and Caco-2 cell surface by lactobacilli. Journal of Medical Microbiology52 (Pt 10), 925–930.
- Lin P.W., Myers L.E.S., Ray L., Song S.-C., Nasr T.R., Berardinelli A.J.et al. (2009) Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radical Biology and Medicine15, 1205–1211.
- Manninen T.J.K., Rinkinen M.L., Beasley S.S. & Saris P.E.J. (2006) Alteration of the canine small-intestinal lactic acid bacterium microbiota by feeding of potential probiotics. Applied and Environment Microbiology72, 6539–6543.
- Mansfield C. (2015) The GI Virome. ECVIM congress: Lisbon, Portugal, 255–256.
- Mar J.S., Nagalingam N.A., Song Y., Onizawa M., Lee J.W. & Lynch S.V. (2014) Amelioration of DSS-induced murine colitis by VSL#3 supplementation is primarily associated with changes in ileal microbiota composition. Gut Microbes5, 494–503.
- Marciňáková M., Simonová M., Strompfová V. & Lauková A. (2006) Oral application of Enterococcus faecium strain EE3 in healthy dogs. Folia Microbiologica (Praha)51, 239–242.
- Marks S.L. & Kather E.J. (2003) Antimicrobial susceptibilities of canine Clostridium difficile and Clostridium perfringens isolates to commonly utilized antimicrobial drugs. Veterinary Microbiology94, 39–45.
- Marks S.L., Kather E.J., Kass P.H. & Melli A.C. (2002) Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. Journal of Veterinary Internal Medicine16, 533–540.
- Martín R., Olivares M., Pérez M., Xaus J., Torre C., Fernández L.et al. (2010) Identification and evaluation of the probiotic potential of lactobacilli isolated from canine milk. Vet J.185, 193–198.
- McCoy S. & Gilliland S.E. (2007) Isolation and characterization of Lactobacillus species having potential for use as probiotic cultures for dogs. Journal of Food Science72, 94–97.
- Medellin-Peña M.J., Wang H., Johnson R., Anand S. & Griffiths M.W. (2007) Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Applied and Environment Microbiology73, 4259–4267.
- Middelbos I.S., Vester Boler B.M., Qu A., White B.A., Swanson K.S., Fahey G.C. (2010) Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE5, e9768.
- Minamoto Y., Otoni C.C., Steelman S.M., Büyükleblebici O., Steiner J.M., Jergens A.E.et al. (2014a) Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes6, 37–41.
- Minamoto Y., Dhanani N., Markel M.E., Steiner J.M. & Suchodolski J.S. (2014b) Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Veterinary Microbiology174, 463–473.
- Miquel S., Martín R., Rossi O., Bermúdez-Humarán L.G., Chatel J.M., Sokol H.et al. (2013) Faecalibacterium prausnitzii and human intestinal health. Current Opinion in Microbiology16, 255–261.
- Momozawa Y., Deffontaine V., Louis E. & Medrano J.F. (2011) Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS ONE6, e16952.
- Murphy T., Chaitman J. & Han E. (2014) Use of fecal transplant in eight dogs with refractory Clostridium perfringens associated diarrhea. Journal of Veterinary Internal Medicine28, 976–1134.
- Oelschlaeger T.A. (2010) Mechanisms of probiotic actions - A review. International Journal of Medical Microbiology300, 57–62.
- ten Oever J. & Netea M.G. (2014) The bacteriome-mycobiome interaction and antifungal host defense. European Journal of Immunology27, 3182–3191.
- Ogué-Bon E., Khoo C., McCartney A.L., Gibson G.R. & Rastall R.A. (2010) In vitro effects of synbiotic fermentation on the canine faecal microbiota. FEMS Microbiology Ecology73, 587–600.
- O'Mahony D., Murphy K.B., MacSharry J., Boileau T., Sunvold G., Reinhart G.et al. (2009) Portrait of a canine probiotic Bifidobacterium-From gut to gut. Veterinary Microbiology139, 106–112.
- Pagnini C., Saeed R., Bamias G., Arseneau K.O., Pizarro T.T. & Cominelli F. (2010) Probiotics promote gut health through stimulation of epithelial innate immunity. Proceedings of the National Academy of Sciences of the United States of America107, 454–459.
- Panasevich M.R., Kerr K.R., Dilger R.N., Fahey G.C., Guérin-Deremaux L., Lynch G.L.et al. (2014) Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. British Journal of Nutrition113, 125–133.
- Perelmuter K., Fraga M. & Zunino P. (2008) In vitro activity of potential probiotic Lactobacillus murinus isolated from the dog. Journal of Applied Microbiology104, 1718–1725.
- Perelmuter K., Fraga M., Delucchi L. & Zunino P. (2011) Safety assessment and enteric colonization ability of a native canine Lactobacillus murinus strain. World Journal of Microbiology & Biotechnology27, 1725–1730.
- Peters I.R., Helps C.R., Calvert E.L., Hall E.J. & Day M.J. (2005) Cytokine mRNA quantification in duodenal mucosa from dogs with chronic enteropathies by real-time reverse transcriptase polymerase chain reaction. Journal of Veterinary Internal Medicine19, 644–653.
- Ren Z., Zhao Z., Wang Y. & Huang K. (2011) Preparation of selenium/zinc-enriched probiotics and their effect on blood selenium and zinc concentrations, antioxidant capacities, and intestinal microflora in canine. Biological Trace Element Research141, 170–183.
- Rérat A., Fiszlewicz M., Giusi A. & Vaugelade P. (1987) Influence of meal frequency on postprandial variations in the production and absorption of volatile fatty acids in the digestive tract of conscious pigs. Journal of Animal Science64, 448–456.
- Rinkinen M., Westermarck E., Salminen S. & Ouwehand A.C. (2003a) Absence of host specificity for in vitro adhesion of probiotic lactic acid bacteria to intestinal mucus. Veterinary Microbiology97, 55–61.
- Rinkinen M., Jalava K., Westermarck E., Salminen S. & Ouwehand A.C. (2003b) Interaction between probiotic lactic acid bacteria and canine enteric pathogens: a risk factor for intestinal Enterococcus faecium colonization?Veterinary Microbiology92, 111–119.
- Roberfroid M., Gibson G.R., Hoyles L., McCartney A.L., Rastall R., Rowland I.et al. (2010) Prebiotic effects: metabolic and health benefits. British Journal of Nutrition104 (Suppl.), S1–S63.
- Rossi G., Pengo G., Caldin M., Piccionello A.P., Steiner J.M., Cohen N.D.et al. (2014) Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS ONE9. doi: 10.1371/journal.pone.0094699.
- Saarela M., Mogensen G., Fondén R., Mättö J. & Mattila-Sandholm T. (2000) Probiotic bacteria: safety, functional and technological properties. Journal of Biotechnology84, 197–215.
- Saez-Lara M.J., Gomez-Llorente C., Plaza-Diaz J. & Gil A. (2015) The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory Bowel disease and other related diseases: a systematic review of randomized human clinical trials. BioMed Research International2015, 505878.
- Sartor R.B. (2006) Mechanisms of disease: pathogenesis of Crohns disease and ulcerative colitis. Nature Clinical Practice. Gastroenterology & Hepatology3, 390–407.
- Sauter S.N., Allenspach K., Gaschen F., Gröne A., Ontsouka E. & Blum J.W. (2005) Cytokine expression in an ex vivo culture system of duodenal samples from dogs with chronic enteropathies: modulation by probiotic bacteria. Domestic Animal Endocrinology29, 605–622.
- Sauter S.N., Benyacoub J., Allenspach K., Gaschen F., Ontsouka E., Reuteler G.et al. (2006) Effects of probiotic bacteria in dogs with food responsive diarrhoea treated with an elimination diet. Journal of Animal Physiology and Animal Nutrition90, 269–277.
- Schmitz S., Garden O.A., Werling D. & Allenspach K. (2012) Gene expression of selected signature cytokines of T cell subsets in duodenal tissues of dogs with and without inflammatory bowel disease. Veterinary Immunology and Immunopathology146, 87–91.
- Schmitz S., Henrich M., Neiger R., Werling D. & Allenspach K. (2013) Comparison of TNFa responses induced by Toll-like receptor ligands and probiotic Enterococcus faecium in whole blood and peripheral blood mononuclear cells of healthy dogs. Veterinary Immunology and Immunopathology Elsevier B.V.; 153, 170–174.
- Schmitz S., Henrich M., Neiger R., Werling D. & Allenspach K. (2014) Stimulation of duodenal biopsies and whole blood from dogs with food-responsive chronic enteropathy and healthy dogs with toll-like receptor ligands and probiotic Enterococcus faecium. Scandinavian Journal of Immunology80, 85–94.
- Schmitz S., Glanemann B., Garden O.A., Brooks H., Chang Y.M., Werling D.et al. (2015a) A prospective, randomized, blinded, placebo-controlled pilot study on the effect of Enterococcus faecium on clinical activity and intestinal gene expression in canine food-responsive chronic enteropathy. Journal of Veterinary Internal Medicine29, 533–543.
- Schmitz S., Werling D. & Allenspach K. (2015b) Effects of ex-vivo and in-vivo treatment with probiotics on the inflammasome in dogs with chronic enteropathy. PLoS ONE10, e0120779.
- Shida K., Nanno M. & Nagata S. (2011) Flexible cytokine production by macrophages and T cells in response to probiotic bacteria: a possible mechanism by which probiotics exert multifunctional immune regulatory activities. Gut Microbes2, 109–114.
- Simpson K.W., Rishniw M., Bellosa M., Liotta J., Lucio A., Baumgart M.et al. (2009) Influence of Enterococcus faecium SF68 probiotic on giardiasis in dogs. Journal of Veterinary Internal Medicine23, 476–481.
- Soo I., Madsen K.L., Tejpar Q., Sydora B.C., Sherbaniuk R., Cinque B.et al. (2008) VSL#3 probiotic upregulates intestinal mucosal alkaline sphingomyelinase and reduces inflammation. Canadian Journal of Gastroenterology22, 237–242.
- Spears J.K., Karr-Lilienthal L.K. & Fahey G.C. (2005) Influence of supplemental high molecular weight pullulan or gamma-cyclodextrin on ileal and total tract nutrient digestibility, fecal characteristics, and microbial populations in the dog. Archives of Animal Nutrition59, 257–270.
- Strompfová V. & Lauková A. (2014) Isolation and characterization of faecal bifidobacteria and lactobacilli isolated from dogs and primates. Anaerobe29, 108–112.
- Strompfová V., Lauková A. & Ouwehand A.C. (2004a) Selection of enterococci for potential canine probiotic additives. Veterinary Microbiology100, 107–114.
- Strompfová V., Lauková A. & Ouwehand A.C. (2004b) Lactobacilli and enterococci–potential probiotics for dogs. Folia Microbiologica (Praha)49, 203–207.
- Strompfová V., Marciňáková M., Simonová M., Bogovič-Matijašić B. & Lauková A. (2006) Application of potential probiotic Lactobacillus fermentum AD1 strain in healthy dogs. Anaerobe12, 75–79.
- Strompfová V., Lauková A. & Gancarčíková S. (2012a) Effectivity of freeze-dried form of Lactobacillus fermentum AD1-CCM7421 in dogs. Folia Microbiologica (Praha)57, 347–350.
- Strompfová V., Plachá I., Čobanová K., Gancarčíková S., Mudroňová D. & Lauková A. (2012b) Experimental addition of Eleutherococcus senticosus and probiotic to the canine diet. Central European Journal of Biology7, 436–447.
- Strompfová V., Pogány Simonová M., Gancarčíková S., Mudroňová D., Farbáková J., Mad'ari A.et al. (2014) Effect of Bifidobacterium animalis B/12 administration in healthy dogs. Anaerobe28, 37–43.
- Suchodolski J.S., Ruaux C.G., Steiner J.M., Fetz K. & Williams D.A. (2005) Assessment of the qualitative variation in bacterial microflora among compartments of the intestinal tract of dogs by use of a molecular fingerprinting technique. American Journal of Veterinary Research66, 1556–1562.
- Suchodolski J.S., Camacho J. & Steiner J.M. (2008) Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiology Ecology66, 567–578.
- Suchodolski J.S., Dowd S.E., Westermarck E., Steiner J.M., Wolcott R.D., Spillmann T.et al. (2009) The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiology9, 210.
- Suchodolski J.S., Xenoulis P.G., Paddock C.G., Steiner J.M. & Jergens A.E. (2010) Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Veterinary Microbiology19, 394–400.
- Suchodolski J.S., Dowd S.E., Wilke V., Steiner J.M. & Jergens A.E. (2012a) 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE7, e39333.
- Suchodolski J.S., Markel M.E., Garcia-Mazcorro J.F., Unterer S., Heilmann R.M., Dowd S.E.et al. (2012b) The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE7, e51907.
- Swanson K.S., Grieshop C.M., Flickinger E.A., Bauer L.L., Chow J., Wolf B.W.et al. (2002) Fructooligosaccharides and Lactobacillus acidophilus modify gut microbial populations, total tract nutrient digestibilities and fecal protein catabolite concentrations in healthy adult dogs. Journal of Nutrition132, 3721–3731.
- Swanson K.S., Dowd S.E., Suchodolski J.S., Middelbos I.S., Vester B.M., Barry K.A.et al. (2011) Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME Journal5, 639–649.
- Tang Y. & Saris P.E.J. (2013) Strain-specific detection of orally administered canine jejunum-dominated Lactobacillus acidophilus LAB20 in dog faeces by real-time PCR targeted to the novel surface layer protein. Letters in Applied Microbiology57, 330–335.
- The NIH HMP Working Group (2009) The NIH Human Microbiome Project. Genome Research19, 2317–2323.
- Thomas C.M. & Versalovic J. (2010) Modulation of signaling pathways in the intestine. Gut Microbes1, 148–163.
- Tsilingiri K., Barbosa T., Penna G., Caprioli F., Sonzogni A., Viale G.et al. (2012) Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut61, 1007–1015.
- Tzortzis G., Baillon M.L., Gibson G.R. & Rastall R.A. (2004) Modulation of anti-pathogenic activity in canine-derived Lactobacillus species by carbohydrate growth substrate. Journal of Applied Microbiology96, 552–559.
- Unterer S., Busch K., Leipig M., Hermanns W., Wolf G., Straubinger R.K.et al. (2014) Endoscopically visualized lesions, histologic findings, and bacterial invasion in the gastrointestinal mucosa of dogs with acute hemorrhagic diarrhea syndrome. Journal of Veterinary Internal Medicine28, 52–58.
- Verlinden A., Hesta M., Hermans J.M. & Janssens G.P.J. (2006) The effects of inulin supplementation of diets with or without hydrolysed protein sources on digestibility, faecal characteristics, haematology and immunoglobulins in dogs. British Journal of Nutrition96, 936–944.
- Viswanathan V.K., Hodges K. & Hecht G. (2009) Enteric infection meets intestinal function: how bacterial pathogens cause diarrhoea. Nature Reviews Microbiology Nature Publishing Group; 7, 110–119.
- Vitetta L., Briskey D., Alford H., Hall S. & Coulson S. (2014) Probiotics, prebiotics and the gastrointestinal tract in health and disease. Inflammopharmacology22, 135–154.
- Weese J.S. (2013) Preliminary clinical and microbiome assessment of stool transplantation in the dog and cat. Journal of Veterinary Internal Medicine27, 604–756.
- Weese J.S. & Anderson M.E.C. (2002) Preliminary evaluation of Lactobacillus rhamnosus strain GG, a potential probiotic in dogs. Canadian Veterinary Journal43, 771–774.
- Weese J.S. & Arroyo L. (2003) Bacteriological evaluation of dog and cat diets that claim to contain probiotics. Canadian Veterinary Journal44, 212–216.
- Weese J.S., Weese H.E., Yuricek L. & Rousseau J. (2004) Oxalate degradation by intestinal lactic acid bacteria in dogs and cats. Veterinary Microbiology101, 161–166.
- White R.S.R.G., Atherly T., Webb C., Hill S., Steiner J.M., Suchodolski J.S.et al. (2015) Effect of VSL#3 probiotic strains on the intestinal microbiota in canine inflammatory bowel disease. Journal of Veterinary Internal Medicine9, 423–483.
- Xenoulis P.G., Palculict B., Allenspach K., Steiner J.M., Van House A.M. & Suchodolski J.S. (2008) Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiology Ecology66, 579–589.
- Zhong Y., Huang J., Tang W., Chen B. & Cai W. (2012) Effects of probiotics, probiotic DNA and the CpG oligodeoxynucleotides on ovalbumin-sensitized Brown-Norway rats via TLR9/NF-κB pathway. FEMS Immunology and Medical Microbiology66, 71–82.
Document information
Published on 09/06/17
Submitted on 09/06/17
Licence: Other
Share this document
Keywords
claim authorship
Are you one of the authors of this document?