| (One intermediate revision by the same user not shown) | |||
| Line 64: | Line 64: | ||
| − | [[Image:draft_Content_147999785-mmc_doc.gif|center|link=File:draft_Content_147999785 | + | [[Image:draft_Content_147999785-mmc_doc.gif|center|link=File:draft_Content_147999785-mmc1.docx]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Conflict of interest== | ==Conflict of interest== | ||
Latest revision as of 14:17, 5 April 2017
Dear Editor,
Parvalbumin is a major allergen of fish allergies, as recognized by most patients with fish allergies.1 The protein is composed of AB, CD, and EF domains and characterized by three EF-hand motifs (helix-loop-helix), two of which (termed CD and EF sites contained in CD and EF domains, respectively) are calcium-binding sites. Ca2+ binding is involved in the structural preservation of parvalbumin; hence, depletion of Ca2+ leads to structural changes.
Regarding the relationship between IgE reactivities and Ca2+ chelation, different results have been obtained. In the case of Baltic cod (Gadus callarias) parvalbumin, its IgE reactivity is slightly reduced (25% reduction) by Ca2+ depletion.2 It is therefore considered that the primary structure is associated with the IgE-binding epitopes of Baltic cod parvalbumin. Indeed, some peptides of it have been identified as IgE-binding epitopes.3 However, the IgE reactivities of parvalbumins have been revealed to be significantly reduced by Ca2+ depletion in common carp (Cyprinus carpio; 30%–90% reduction) and Pacific mackerel (Scomber japonicus; 60%–100% reduction). 4 and 5 Therefore, these data indicate the significance of conformational IgE-binding epitopes. 4 and 5 Indeed, some amino acid residues of common carp parvalbumin have been proposed to comprise conformational IgE-binding epitopes.6 Although these differing results have been revealed, parvalbumin cross-reacts with various types of fish, 1, 7 and 8 indicating that parvalbumins have common IgE-binding epitopes. To confirm whether conformational IgE-binding epitopes exist in a stereoscopic conformation maintained by Ca2+ binding, this study assessed the influence of Ca2+ on the stereoscopic conformation and IgE reactivities of parvalbumins.
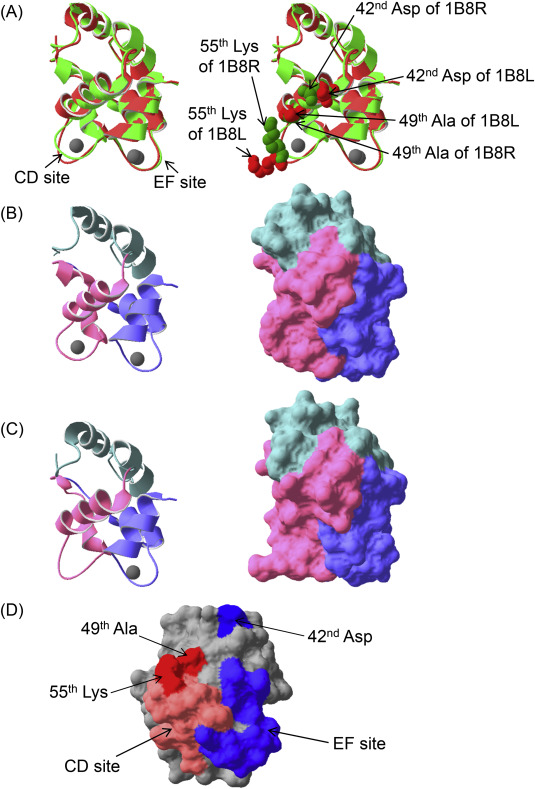
First, the common carp parvalbumin 1B8R,9 which can bind two Ca2+ molecules in the CD and EF sites, was compared with the CD site-mutated parvalbumin 1B8L,9 which binds one Ca2+ molecule only in the EF site (Fig. 1A). The loop in CD site of parvalbumin 1B8L was narrower than that of parvalbumin 1B8R (Fig. 1B,C, left). Regarding their molecular surfaces, the CD domain of parvalbumin 1B8L is dramatically different from that of parvalbumin 1B8R (Fig. 1B,C, right). Therefore, if IgE-binding epitopes exist in a conformational structure, depletion of Ca2+ could affect IgE reactivities. In our past report,5 we described the production of single CD or EF site-mutated pacific mackerel parvalbumins, and a CD and EF sites co-mutated parvalbumin. These mutated sites lost Ca2+-binding abilities. Both single mutated parvalbumins showed moderate reductions of IgE reactivities, and the co-mutated parvalbumin lost almost all IgE reactivities. Therefore, IgE reactivities were affected by conformational changes of not only the CD site but also the EF site.
|
|
|
Fig. 1. Comparison of two recombinant common carp (Cyprinus carpio) parvalbumins and candidates of fish-specific residues for IgE-bindings. Superposition of ribbon diagrams of carp parvalbumin 1B8R, which can bind two Ca2+ molecules in the CD and EF sites [light green, Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) accession ID 1B8R], and the CD site-mutated carp parvalbumin 1B8L, which bind only one Ca2+ molecule in the EF site (light red, Protein Data Bank accession ID 1B8L) (A). A ribbon diagram of parvalbumin 1B8R and 1B8L (B and C, left, respectively). Molecular surfaces of parvalbumin 1B8R and 1B8L (B and C, right, respectively). Stereoscopic structures of both carp parvalbumins were analyzed by X-ray diffraction.9 Gray spherical objects indicate Ca2+ ions in (A), (B), and (C). Mint green, light pink, and light blue regions denote the AB, CD, and EF domains, respectively, in (B) and (C). Candidates of fish-specific crucial residues for allergen-antibody interactions on the molecular surface of Pacific mackerel parvalbumin (D). Light orange and light blue in (D) indicate CD or EF sites, respectively. Dark red and dark blue in (D) indicate candidates associated with the CD and EF sites, respectively. These candidates are also shown in right side of (A). The structure of Pacific mackerel parvalbumin was constructed based on the common carp parvalbumin revealed by X-ray diffraction (Protein Data Bank accession ID 1CDP) using Swiss-PdbViewer (http://spdbv.vital-it.ch/) and was optimized using TINKER molecular modeling software (http://dasher.wustl.edu/tinker/). All figures were produced using Swiss-PdbViewer. |
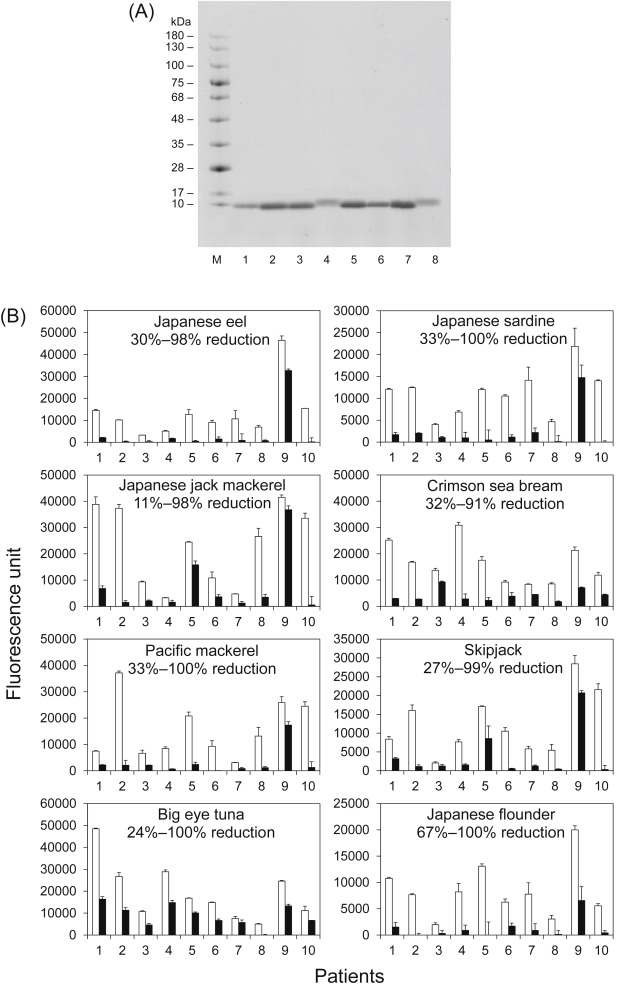
Next, parvalbumins were purified from eight fish species (Fig. 2A; see Supplementary Methods and Supplementary Fig. 1 for details). Further, their IgE reactivities in the absence or presence of ethylene glycol tetraacetic acid (EGTA) as a Ca2+ chelating agent were assessed by enzyme-linked immunosorbent assay (ELISA), as previously described (see Supplementary Methods for details).5 In the absence of EGTA, parvalbumin exists in a Ca2+-bound form. Contrarily, parvalbumin releases Ca2+ in the presence of EGTA. Except for patient 9, the IgE reactivities of parvalbumins with EGTA from all fish species were remarkably reduced compared with that in the absence of EGTA (Fig. 2B) as follows: average reduction: Japanese eel, 83%; Japanese sardine, 83%; Japanese jack mackerel, 68%; crimson sea bream, 72%; Pacific mackerel, 81%; skipjack, 72%; big eye tuna, 53%; and Japanese flounder, 88%. The pooled control serum did not react to all samples (data not shown). These findings demonstrated the significance of Ca2+ for IgE-bindings. These data strongly suggest that the IgE-binding capacity of parvalbumin from most types of fish other than the aforementioned fish may also mainly depend on the conformational structure of parvalbumin. In addition, the reduction in IgE reactivities to parvalbumins from all fish species were lower in the case of patient 9, when EGTA was present as opposed to cases of the other patients. Presumably, patient 9 recognizes not only the conformational IgE-binding epitope(s) but also the linear IgE-binding epitope. Moreover, the reductions in IgE reactivities to big eye tuna parvalbumin were slightly lower in all patients. Therefore, its parvalbumin may possess a species-specific linear IgE-binding epitope in addition to conformational epitope(s).
|
|
|
Fig. 2. Analysis of SDS-PAGE and IgE reactivities of purified parvalbumin. SDS-PAGE of purified parvalbumins (A). M, protein marker; 1, Japanese eel; 2, Japanese sardine; 3, Japanese jack mackerel; 4, crimson sea bream; 5, Pacific mackerel; 6, skipjack; 7. big eye tuna; and 8, Japanese flounder. Analysis of the IgE reactivity of parvalbumins obtained from eight types of fish in the absence (open bars) and presence (closed bars) of ethylene glycol tetraacetic acid by enzyme-linked immunosorbent assay (B). The reduction in IgE reactivities to parvalbumins is shown in each graph as %. |
Supplementary Figure 2 shows alignment of amino acid sequences of fish and mammalian parvalbumin. Identities among fish were 67%–85%, but those between human and fish were only 44%–48%. The CD sites of fish parvalbumins had several differences from mammalian parvalbumin. However, these characteristics were similar to the residues of fish parvalbumin. Because the mutation of CD site led to a moderate reduction in IgE reactivity,5 sequentially separated residues important for IgE-binding must exist. For example, the 49th Ser/Ala and its small side chain located near the CD site (Fig. 1A,D) have different characteristics from the 49th residues of mammalian parvalbumins (Supplementary Fig. 2). In addition, the 55th Lys in the CD site (Fig. 1A,D) was conserved in fish, but included a different residue from the 55th residue of the mammalian parvalbumin, which is not charged (Supplementary Fig. 2). Lysine is positively charged and can form ion bonds, suggesting it is a crucial fish-specific residue for IgE-binding. Hence, these residues may be a part of an IgE-binding epitope. In contrast, the primary structures of the fish and mammalian EF site were similar. Because the EF site-mutated parvalbumin showed a moderate reduction of IgE reactivities,5 a crucial residue for IgE binding is expected to be sterically located near the EF site. For example, the 42nd Asp, which is located at the periphery of the EF site (Fig. 1A,D), is fish-specific (Supplementary Fig. 2) and may be related to the ionic interactions in IgE-binding.
Previous findings4, 5 and 6 and our results strongly suggest that IgE-binding epitopes present in the stereoscopic conformation of parvalbumin are highly conserved in many fish species. Fish parvalbumin is a highly cross-reactive panallergen.1, 7 and 8 Therefore, it is hoped that conformational IgE-binding epitopes exhibiting cross-reactivities are revealed in fish species other than common carp.
Acknowledgments
The authors thank Drs. H. Ohsuna and Z. Ikezawa (Department of Dermatology, Yokohama City University) for providing patient sera. This study was partly supported by Japan Society for the Promotion of Science KAKENHI Grant Number 25750023.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Conflict of interest
The authors have no conflict of interest to declare.
References
- 1 Y. Hamada, H. Tanaka, S. Ishizaki, M. Ishida, Y. Nagashima, K. Shiomi; Purification, reactivity with IgE and cDNA cloning of parvalbumin as the major allergen of mackerels; Food Chem Toxicol, 41 (2003), pp. 1149–1156
- 2 J. Apold, S. Elsayed; The effect of amino acid modification and polymerization on the immunochemical reactivity of cod allergen M; Mol Immunol, 16 (1979), pp. 559–664
- 3 S. Elsayed, J. Apold; Immunochemical analysis of cod fish allergen M. locations of the immunoglobulin binding sites as demonstrated by the native and synthetic peptides; Allergy, 38 (1983), pp. 449–459
- 4 I. Swoboda, A. Bugajska-Schretter, P. Verdino, W. Keller, W.R. Sperr, P. Valent, et al.; Recombinant carp parvalbumin, the major cross-reactive fish allergen: a tool for diagnosis and therapy of fish allergy; J Immunol, 168 (2002), pp. 4576–4584
- 5 S. Tomura, S. Ishizaki, Y. Nagashima, K. Shiomi; Reduction in the IgE reactivity of Pacific mackerel parvalbumin by mutations at Ca2+-binding sites; Fish. Sci, 74 (2008), pp. 411–417
- 6 E. Untersmayr, K. Szalai, A.B. Riemer, W. Hemmer, I. Swoboda, B. Hantusch, et al.; Mimotopes identify conformational epitopes on parvalbumin, the major fish allergen; Mol Immunol, 43 (2006), pp. 1454–1461
- 7 Y. Hamada, H. Tanaka, A. Sato, S. Ishizaki, Y. Nagashima, K. Shiomi; Expression and evaluation of IgE-binding capacity of recombinant Pacific mackerel parvalbumin; Allergol Int, 53 (2004), pp. 271–278
- 8 Y. Kobayashi, J. Huge, S. Imamura, N. Hamada-Sato; Study of the cross-reactivity of fish allergens based on a questionnaire and blood testing; Allergol Int, 65 (2016), pp. 272–279
- 9 M.S. Cates, M.B. Berry, E.L. Ho, Q. Li, J.D. Potter, G.N. Phillips Jr.; Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin; Structure, 7 (1999), pp. 1269–1278
Document information
Published on 05/04/17
Licence: Other
Share this document
claim authorship
Are you one of the authors of this document?